Abstract
The Bloom protein (BLM) and Topoisomerase IIIα are found in association with proteins of the Fanconi anemia (FA) pathway, a disorder manifesting increased cellular sensitivity to DNA crosslinking agents. In order to determine if the association reflects a functional interaction for the maintenance of genome stability, we have analyzed the effects of siRNA-mediated depletion of the proteins in human cells. Depletion of Topoisomerase IIIα or BLM leads to increased radial formation, as is seen in FA. BLM and Topoisomerase IIIα are epistatic to the FA pathway for suppression of radial formation in response to DNA interstrand crosslinks since depletion of either of them in FA cells does not increase radial formation. Depletion of Topoisomerase IIIα or BLM also causes an increase in sister chromatid exchanges, as is seen in Bloom syndrome cells. Human Fanconi anemia cells, however, do not demonstrate increased sister chromatid exchanges, separating this response from radial formation. Primary cell lines from mice defective in both Blm and Fancd2 have the same interstrand crosslink-induced genome instability as cells from mice deficient in the Fancd2 protein alone. These observations demonstrate that the association of BLM and Topoisomerase IIIα with Fanconi proteins is a functional one, delineating a BLM-Topoisomerase IIIα-Fanconi pathway that is critical for suppression of chromosome radial formation.
Key Words: Bloom protein, Fanconi anemia, Genome stability, Sister chromatid exchange, Topoisomerase IIIα
Introduction
Bloom syndrome (BS) is a rare recessive disorder characterized by growth retardation, immunodeficiency, photosensitivity, and an increased incidence of cancers including leukemias, lymphomas, and carcinomas [German, 1995; Moses, 2001; Hickson, 2003]. Cells from BS patients manifest chromosomal aberrations, including sister chromatid exchanges (SCEs) [Chaganti et al., 1974] which are used for clinical diagnosis, and increased radials. The elevated SCEs reflect increased interchromosomal homologous recombination (HR), apparently in S-phase [German, 1995; Sonoda et al., 1999; Gaymes et al., 2002]. The defect in BS is in the Bloom protein (BLM), a member of the conserved RecQ helicase family, of which there are five recognized paralogs in humans [Wu and Hickson, 2006]. RecQ proteins appear to regulate recombination, thus enhancing global genome stability, and are involved in the response to a variety of DNA adducts [Karow et al., 2000; Chester et al., 2006; Hanada and Hickson, 2007]. BS cells also show abnormal replication intermediates with prolonged S-phase delay after DNA damage [Davies et al., 2004] and fail to re-start replication efficiently after blockade [Davies et al., 2007]. Thus it appears a deficiency in a RecQ enzyme leads to an increase in recombination, possibly as the result of a replication defect, leading to SCE formation [Hanada et al., 1997; Hanada and Hickson, 2007].
While deficiency of the RecQ protein is not lethal in prokaryotes, disruption of the Blm gene in mice is lethal in utero [Chester et al., 1998]. Hypomorphic mutations (blmm3/m3) in mice permit viability, but the mice show an increased susceptibility to lymphomas, sarcomas, and carcinomas, which mimics the cancer spectrum seen in BS patients. The development of cancers correlates inversely with the level of BLM [Luo et al., 2000; McDaniel et al., 2003].
BLM has a conserved association with the type IA topoisomerase, Topoisomerase IIIα (Topo IIIα). As with BLM, a defect in Topo IIIα leads to embryonic lethality in mice [Li and Wang, 1998]. In budding yeast, deficiency of Top3 leads to a hyper-recombination phenotype, and top3 mutants are sensitive to DNA damaging agents[Chakraverty et al., 2001; Ira et al., 2003]. The physical interaction of Sgs1, and Top3 [Bennett et al., 2000] reflects a functional interdependence; mutations in SGS1 can occur as suppressor mutations for top3 mutants [Gangloff et al., 1994]. Along with a third protein, Rmi1, Sgs1 and Top3 appear to maintain genome stability in S-phase [Schmidt and Kolodner, 2004]. The mammalian homolog of Rmi1, BLAP75 or hRMI1, and RMI2 associate with Topo IIIα to stimulate activity of BLM on HR intermediates [Wu and Hickson, 2003; Chang et al., 2005; Mullen et al., 2005; Wu et al., 2006; Bussen et al., 2007; Singh et al., 2008; Xu et al., 2008]. Studies on Topo IIIα-deficient mammalian cells are lacking, due to cell lethality, but a conditional disruption has been used in chicken cells to demonstrate sensitivity to DNA damage [Otsuki et al., 2008]. Overall it appears that Topo IIIα and BLM affect genome stability in a common pathway by modulation of HR.
Fanconi anemia (FA) is a rare recessive disorder resulting from a deficiency in any of at least 13 proteins [D'Andrea and Grompe, 2003; Wang, 2007]. It manifests growth abnormalities, deficiencies in all blood cell lineages, and an increased risk of malignancies [Moses, 2001; D'Andrea and Grompe, 2003]. FA cells show increased cellular sensitivity to agents that form DNA interstrand crosslinks (ICLs), as manifested by chromosomal breaks and radials [Schroeder et al., 1964]. The FA proteins have been shown to interact with BRCA1 [Garcia-Higuera et al., 2001; Folias et al., 2002], and the BRCA2 gene has been identified as the FANCD1 gene [Howlett et al., 2002]. In addition, two BRCA-associated proteins, PALB2 and BRIP1 (BACH1), are FANCN and FANCJ, respectively [Bridge et al., 2005; Levitus et al., 2005; Reid et al., 2007]. Thus the FA/BRCA proteins comprise a DNA damage response network required for genome stability.
The BLM/Topo IIIα and FA/BRCA pathways each act to maintain genome stability as outlined above. A link between the FA pathway and the BS pathway is suggested by the observation that five FA proteins, FANCA, C, E, F and G, associate with BLM as well as Topoisomerase IIIα and RPA in a complex termed BRAFT [Meetei et al., 2003; reviewed in Wang, 2007]. It has also been noted that phosphorylation of BLM, a normal response to DNA damage, appears to be abrogated in FA cells, suggesting an intact FA core complex is required for the modification [Pichierri et al., 2004]. Furthermore, FANCC and BLM co-localize following DNA damage [Hirano et al., 2005]. These observations raise the question as to whether BLM and Topo IIIα act functionally in the FA pathway for genome stability, and also whether FANC proteins participate in suppression of SCEs in human cells. Findings differ regarding SCE formation in FA cells, with reports of normal levels in human FA-A and FA-D1 cells [Godthelp et al., 2006] or elevated levels [Hirano et al., 2005] in FA chicken cells. With BRCA2 (FANCD1) mutations it is reported that SCE formation decreases in rodent cells [Tutt et al., 2001].
We have used normal human cells with siRNA-mediated depletion of Topo IIIα, BLM, or FA, as well as FA and BS patient cells, to investigate whether the BLM and FA pathways functionally interact. Chromosomal radial formation was used as the index for genome stability. We find that normal human cells depleted for BLM or Topo IIIα display a decreased survival in response to mitomycin C (MMC), in addition to the expected increase in SCEs and radial formation. In contrast, depletion of BLM or Topo IIIα in FA cells does not cause an increase in radials, indicating a common response pathway. In the converse test, depletion of FANCA in a BS cell line does not increase radials, again demonstrating epistasis. Cells derived from Fancd2–/–/Blmm3/m3 mice or Fancd2–/– mice show the same sensitivity to MMC, again indicating epistasis. Five complementation groups of human FA cells show normal levels of SCE formation, and depletion of BLM or Topo IIIα in FA cells increases SCE formation, distinguishing the FA pathway from BS for SCE regulation. Monoubiquitination of FANCD2 remains normal in human cells depleted of Topo IIIα, as it does in BS cells. Our results support the conclusion that BLM and Topo IIIα function exclusively in the FA response to ICL formation, establishing a BLM-Topo IIIα-FA pathway and demonstrating a functional basis for the BRAFT complex. Our results support a model wherein BLM-Topo IIIα act in the FA pathway for genome stability after ICL formation, but in a separate pathway for suppression of SCEs.
Materials and Methods
Animal Husbandry
The construction of the Fancd2–/+ mouse in C57BL/6J background has been described [Houghtaling et al., 2003]. Mice heterozygous for the Blmm3 allele [Luo et al., 2000] in a 129S5 background were crossed with the Fancd2–/+ mice. The Fancd2 and Blm mice were bred to produce mice heterozygous for both mutations. Double heterozygous mice were then bred to produce mice homozygous for both mutations in a mixed C57BL/6J×129S5 background.
Quantitative PCR (qRT-PCR)
Total RNA was extracted using the RNeasy Mini Kit (Qiagen). 1 μg RNA was used to synthesize cDNA with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real Time quantitative PCR was performed using the iCyler iQ Detection System (Biorad) with 10 ng of starting cDNA material and the TaqMan Gene Expression Assays specific to primers Topo IIα and Topo IIIα and the endogenous control for the β-actin gene (Applied Biosystems). Samples were tested in triplicate and mRNA concentrations for each sample were calculated using the comparative Ct method [Livak and Schmittgen, 2001].
Mouse Genotyping
The Fancd2 allele was genotyped as previously described [Houghtaling et al., 2003]. The Blm mice were genotyped by Southern blot, as previously described [McDaniel et al., 2003]. 10 μg of genomic DNA was digested with BamHI, and probed using a 1.6-kb probe that spanned exon 4. Blots were probed overnight, and washed at a high stringency (65°C for 30 min in 2× SSC with 0.5% SDS followed by 30 min in 1× SSC). The mutant allele gives a band of 6.0 kb, and the wild-type allele gives a band of 6.6 kb.
Cell Lines and Culture
Transformed fibroblasts [Bruun et al., 2003] were cultured in α-MEM medium (Mediatech) supplemented with 5% fetal bovine serum (Hyclone), 5% calf serum (Hyclone) and 0.05 mg/ml gentamicin (Gibco) in a humidified incubator with 5% CO2 at 37°C. Normal cell line GM639, the FANCA line GM6914, and the Bloom syndrome cell line GM08505 were obtained from the NIGMS Human Genetic Cell Repository. Transformed Fanconi anemia cell lines PD331 (FANCC), VU423 (FANCD1) and PD20 (FANCD2), as well as primary Fanconi anemia cell lines PD438F (FANCC) and PD829 (FANCG), were provided by the OHSU Fanconi Anemia Cell Repository. The retrovirally corrected controls for GM08505, GM6914, PD438F, PD331, and PD829 were functionally complemented with a pMMP retrovirus vector containing a full-length cDNA for the appropriate complementation group. Cell lines PSNF5 and PSNG13 have been previously described [Davies et al., 2004]. Mouse primary fibroblasts were derived from the adult ears for each of the genotypes of interest. The ears were soaked in 70% ethanol, rinsed several times with PBS, minced in RPMI medium (Gibco) containing collagenase (Gibco) and incubated at 37°C for 3 h. The collagenase-containing medium was then replaced with DMEM medium containing 20% fetal calf serum (Hyclone) and the cells were grown at 37°C in a humidified incubator with 5% CO2.
Design of the siRNA Oligonucleotides
The siRNA duplex specific for BLM was designed as previously described [Bruun et al., 2003]. A 21-mer beginning with an AA repeat was selected and specificity to BLM was confirmed by a BLAST analysis. The duplex was synthesized with a 3′-dTT overhang by Dharmacon Research (Lafayette, CO). The sequence is: UCCCGGGAUACUGCUCUCA.dT.dT. For controls, a non-functioning duplex with the following sequence was used: AAC UUU UGC AAA GCG GAG CCA UU. The FANCA and BRCA1-specific duplexes have been described previously [Bruun et al., 2003]. siGENOME SMART pools for Topo IIIα were from Dharmacon: GAAACUAUCUGGAUGUGUAUU, CCACAAAGAUGGUAUCGUAUU, CCAGAAAUCUUCCACAGAAUU, and GAAAAGUCUGACCAAGCUUU. For Topo IIα they were: AAACAGACAUGGAUGGAUAUU, CAAACUACAUUGGCAUUUAUU, GAAAGAGUCCAUCAGAUUUUU, and CGAAAGGAAUGGUUAACUAUU.
Transfection
Transfections were performed as previously described [Bruun et al., 2003]. Final volumes of 1 ml for a T25 flask and 3.2 ml for a 100-mm dish were used. Controls were transfected with either a non-functional siRNA duplex or mock transfected. Transfection efficiency for the BLM siRNA was monitored using the FAM Silencer siRNA Labeling Kit (Ambion). At 4 and 24 h, cells were fixed and counter-stained with DAPI using the SlowFade Light Antifade Kit with DAPI (Molecular Probes). Cells were then imaged using a Leica DRMXA fluorescent microscope. Transfection efficiency was calculated by number of cells with at least one labeled siRNA divided by the total number of DAPI stained cells. For GM639 cells, efficiency was 60% at 4 h and 88% at 24 h for both control and BLM siRNA and 67% for BLM and control siRNA in GM6914.
Cell Survival Assay
Twenty-four hours following transfection, cells were plated on 100-mm dishes at 300 cells per dish and treated with MMC in duplicate. Cells were allowed to grow for 10 days in MMC, and then fixed in a solution of 50% MeOH, 1% freshly prepared methylene blue (Sigma). Colonies were counted, and standard errors of the means, denoted by bars, were derived from the duplicate data sets.
Immunoblotting
Cells were transfected on 100-mm dishes as described above, using a 3.2-ml final solution volume, and were washed with PBS, trypsinized, pelleted, and frozen at −80°C at the indicated time points. The untreated control was mock-transfected and harvested at 48 h. Cell lysates were prepared as previously described [Bruun et al., 2003]. 50 μg whole protein extract from each lysate was run on a 7.5% acrylamide gel. Blots for BLM were then transferred to a nitrocellulose membrane (Osmonics), and FANCA, BRCA1 and FANCD2 blots transferred to Immobilon-P PVDF (Millipore). Membranes were blocked overnight in TBST (TBS plus 0.1% Tween) plus 5% dry milk. For BLM blots, the membrane was probed with anti-BLM rabbit polyclonal antibody (Abcam) at a 1:1,000 dilution in TBST with milk. BRCA1 blots were probed with anti-BRCA1 mouse monoclonal antibody (Ab-4, Oncogene) at a 1:200 dilution in TBST with milk. FANCA blots were probed with anti-FANCA mouse monoclonal antibody (5G9, a gift from Maureen Hoatlin) at a 1:250 dilution in TBST with milk. The β-tubulin blots were probed with a rabbit polyclonal antibody (H-235, Santa Cruz) at a 1:3,000 dilution in TBST and visualized with HRP-conjugated secondary antibodies and enhanced chemiluminescence.
Chromosome Stability
For sister chromatid exchange (SCE) analysis, cells were allowed to go through two rounds of replication in 25 μg/ml BrdU, 24 h post-transfection. Clastogen-induced SCEs were elicited by a 4-h pulse of 20 ng/ml MMC 20 h prior to harvest. Cells were harvested following a 3-h exposure to 0.05 μg/ml colcemid (Gibco), treated with a solution of 1:3, 5% fetal calf serum:0.075 M KCl, and were fixed in 3:1 methanol:acetic acid. Cells were then dropped onto slides and stained for 5 min in 0.01% Acridine Orange. Following staining, the slides were rinsed with deionized water and treated with Sorenson Buffer (pH 6.8) (1:1 volume of 0.06 M Na2HPO4 and 0.06 M KH2PO4). After treatment, the slides were exposed to UV light for 12 min, and then visualized using a FITC filter. 20–25 metaphases from each culture were scored for chromosome count and the number of SCEs. The SCE rate was calculated as number of SCEs per chromosome.
For chromosome breakage studies, cells were treated with MMC (5–40 ng/ml) or diepoxybutane (DEB) (50–150 ng/ml) 24 h after transfection or mock transfection. Cells were treated with a dose of MMC that produced a 5–10% decrease in survival. Following 48 h incubation with the clastogens, cells were harvested as described [Bruun et al., 2003]. Slides were stained with Wright's stain, and 50 metaphases from each culture were scored for radial formation. For murine cells, 10 ng/ml MMC was used for radial studies. The number of cells with radials is defined as a cell with one or more radial.
Cell Cycle Analysis
Cells were transfected in 100-mm dishes as described above. Duplicate plates were transfected to assure appropriate cell numbers. 24 h post transfection, cells were treated with the indicated amount of MMC. After 48 h in the dark, cells were harvested and fixed in ice-cold 70% EtOH. Before analysis, cells were harvested at 200 g for 5 min, and the ethanol was removed. The cells were then incubated for 2 h at 37°C in a DNA extraction buffer of 0.2 M phosphate citrate buffer, pH 7.8. The cells were then harvested by centrifugation for 10 min at 1,500 g, and the supernatant was removed. Cells were stained in a 20 μg/ml solution of propidium iodide in PBS with 0.1% Triton X-100 and 200 μg/ml DNase-free RNase, and were then incubated for 30 min at room temperature in the dark.
The stained cells were sorted on a Becton Dickinson FACSCalibur machine, using CellQuest software for analysis. Approximately 20,000 cells per sample were analyzed.
Results
Depletion of Topo IIIα or BLM Increases Radial and SCE Formation
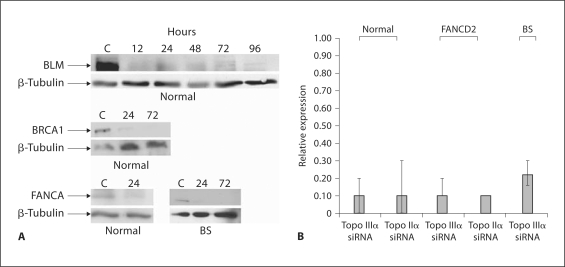
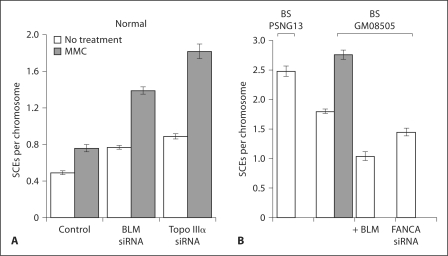
In order to analyze the effects of loss of BLM or Topo IIIα on the ICL response in human fibroblasts, we used siRNA-mediated depletion. Depletion of target proteins or mRNA was verified by immunoblot or qRT-PCR analysis (fig. 1A, B). Protein levels decreased within 12 h and remained depleted for approximately 100 h, allowing testing of chromosome stability and cell survival. A hallmark of BS cells is increased spontaneous SCE formation compared to normal cells, and BLM depletion in normal cells produced increased SCE formation like that in BS cells (fig. 2A). Depletion of Topo IIIα in normal fibroblasts mimicked the findings seen with BLM depletion. MMC treatment increased SCE formation in all cases (fig. 2A). For comparison, BS cells were corrected with retroviral transformation with the BLM gene, resulting in a reduction in SCE formation (fig. 2B).
Fig. 1.
Depletions in normal transformed human fibroblasts (GM639) and BS cells (GM08505). A Immunoblot of BLM, BRCA1 and FANCA depletions. Cells were transfected with siRNA, and harvested at the indicated times. β-Tubulin is shown to indicate loading. Cell lines are indicated under the blots. B Depletion of Topo IIα and Topo IIIα in normal (GM639), BS (GM08505) and FANCD2 (PD20) cells determined by qRT-PCR. Relative expression is in comparison to the non-depleted cell line.
Fig. 2.
SCE formation in normal (A) and BS (B) cells. Normal (GM639) or BS cells (PSNG13 and GM08505) were treated with MMC as described in Materials and Methods and analyzed for SCE formation, with the indicated depletions. SCE are given per chromosome. + BLM indicates a complementation Bloom line, as described in Materials and Methods. The control data sets are from mock-transfected cells. Error bars represent the standard error of the mean.
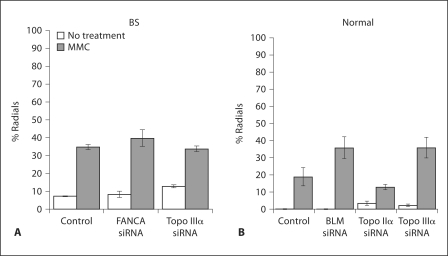
Elevated levels of radial chromosomes are observed following ICL damage in BS cells (fig. 3A). Depletion of BLM in normal fibroblasts produced increased chromosome radials compared to normal cells (fig. 3B), authenticating the depletion. An increased number of radials was also seen in normal cells with MMC following Topo IIIα depletion (fig. 3B). However, depletion of Topo IIIα in BS cells did not increase radials (fig. 3A), indicating that BLM and Topo IIIα are epistatic for suppression of radial formation. The findings demonstrate that the association of BLM and Topo IIIα is a functional relationship for response to ICL formation. The observations with radials are specific, since depletion of topoisomerase IIα, verified by qRT-PCR (fig. 1B), does not promote radial formation following MMC in normal cells (fig. 3B).
Fig. 3.
Chromosomal radial formation. A BS (GM08505) or B normal cells (GM639), were analyzed for radial formation as described in Materials and Methods with MMC (40 ng/ml) and the depletions indicated; the control data are from mock-transfected cells. Percent radials is the percent of cells with one or more radials. Error bars represent standard errors of the mean. No radials were observed in untreated normal cells.
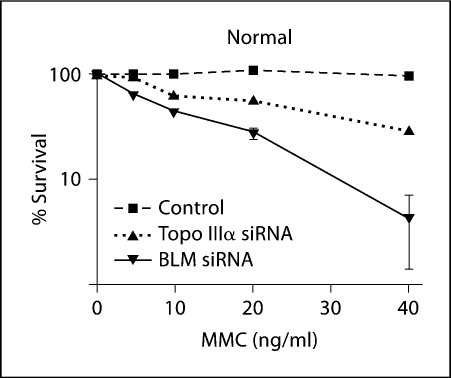
To provide another measure of sensitivity to ICLs in normal human cells depleted of BLM or Topo IIIα, cell survival was determined. Following depletion of BLM or Topo IIIα, a normal fibroblast line showed decreased survival following MMC treatment, relative to untreated cells (fig. 4). Thus by the indices of SCE formation, radial formation and cell survival, depletion of BLM or Topo IIIα produced decreased genome stability, correlating with increased cell sensitivity to DNA crosslinking agents.
Fig. 4.
Survival of normal (GM639) cells depleted for BLM or Topo IIIα after treatment with varying amounts of MMC. Survival was determined by colony formation, normalized to a mock-treated control plate. Error bars represent the standard error of the mean.
Depletion of Topo IIIα Causes Cells to Arrest with 4C DNA Content
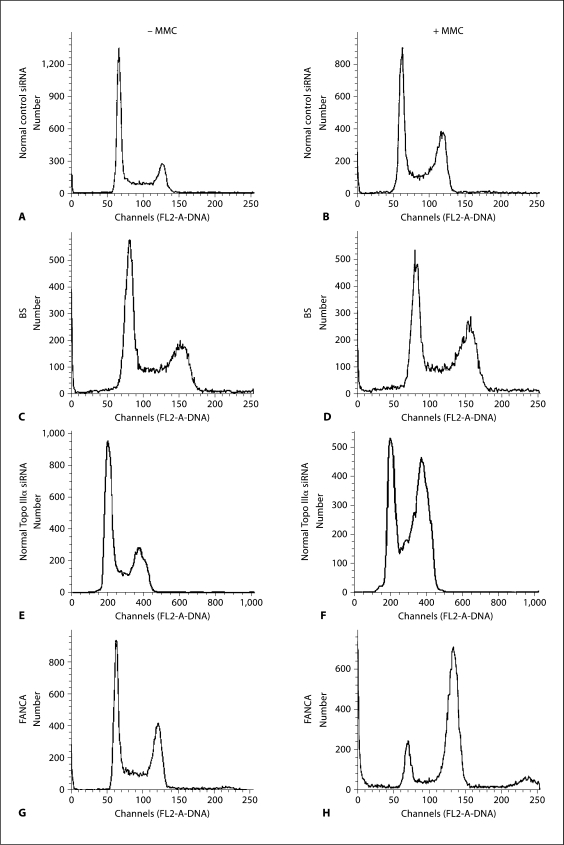
BS cells manifest a G2 DNA increase, compared to normal cells, reflecting a transient arrest at entry into mitosis, as a result of checkpoint activation [Chester et al., 1998; Hickson, 2003; Davies et al., 2004] (fig. 5A–D). We therefore tested whether cells depleted for Topo IIIα manifest a similar profile. Analysis of a normal cell line depleted for Topo IIIα, showed G2 delay (fig. 5E, F), as do BS cells, following MMC treatment. FANCA cells show a similar profile (fig. 5G, H), as has been noted [Wang, 2007]. One interpretation of the delay would be that in the absence of BLM or Topo IIIα, a late S-phase or G2 checkpoint mechanism is activated, due to increased strand breaks persisting from replication, leading to fork collapse, as may be the case in BS cells [Davies et al., 2004].
Fig. 5.
Cell cycle analysis with BLM or Topo IIIα depletion. Cells with or without MMC (80 ng/ml) were analyzed for DNA content. Cells are GM639 (normal) with control siRNA (A, B), GM08505 (BS) (C, D), GM639 with Topo IIIα siRNA (E, F), and GM6914 (FANCA) (G, H).
Topo IIIα and BLM Act Epistatically with FA for Suppression of Radials
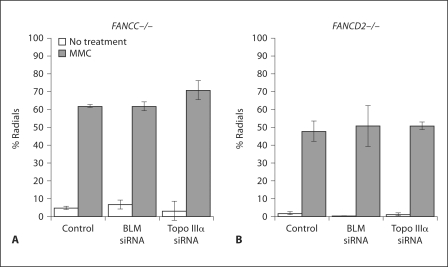
Since a deficiency in BLM, Topo IIIα or FANC proteins decreases genome stability in a similar manner, and causes a similar G2 delay, and because of the observed physical interaction between these proteins, we tested whether FANC proteins, BLM and Topo IIIα function in the same pathway in response to ICLs. In contrast to the above results, depleting BLM or Topo IIIα in FANCD2-deficient or FANCC-deficient cell lines did not increase the incidence of chromosomal instability as measured by radial formation following MMC treatment (fig. 6). The same was found for a FANCA cell line (data not shown). The converse test, depletion of FANCA in BS cells, did not increase radial formation in response to MMC (fig. 3A), supporting the conclusion that BLM and Topo IIIα are epistatic to the FA pathway in response to ICL formation and do not function in additional pathways for ICL response, at least as indexed by radial formation. As expected, depletion of Topo IIα did not increase radial formation in FA cells, since the depletion had no effect in normal cells. The epistasis of BLM and the FA pathway was validated by cell survival following MMC depletion of BLM in a FANCD2 cell line; there was no decrease in survival after MMC treatment compared to the same line non-depleted (data not shown).
Fig. 6.
Radial formation in FA cells depleted for BLM or Topo IIIα. FANCC (PD331) (A) or FANCD2 (PD20) (B) cells were mock treated or depleted for the indicated protein. Radials were analyzed with or without MMC treatment (40 ng/ml) as in Materials and Methods. Error bars represent standard error of the mean.
We have previously reported that depletion of BRCA1 in FA cells increases radial formation, indicating BRCA1 must act in at least one additional pathway for genome stability [Bruun et al., 2003]. The same is true for BRCA1 depletion in BS or Topo IIIα-depleted cells, as would be expected if the latter two proteins are epistatic with FA (data not shown).
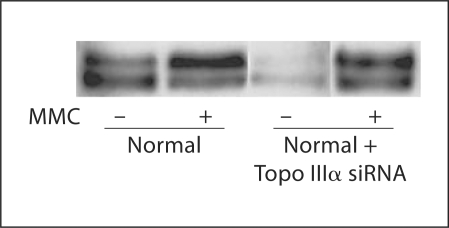
FANCD2 protein is modified following DNA damage and in S-phase by monoubiquitination (FANCD2-Ub) [Garcia-Higuera et al., 2001; Timmers et al., 2001; D'Andrea and Grompe, 2003]. BS cells manifest normal FANCD2-Ub formation [Pichierri et al., 2004]. Therefore, depletion of Topo IIIα should not affect the modification of FANCD2, as was the case (fig. 7). Thus, while depletion of BLM or Topo IIIα causes increased radials and decreased survival in response to ICL formation, this is not due to a deficiency of FANCD2-Ub formation.
Fig. 7.
Monoubiquitination of FANCD2 with depletion of Topo IIIα. Lanes 1 and 2, normal (GM639) cells with control siRNA; 3 and 4 with Topo IIIα siRNA. Cells were harvested 24 h after MMC treatment, as described in Materials and Methods.
FA Does Not Regulate SCE Formation
Due to differing reports regarding SCE formation in FA cells from model rodent or chicken cells and in human cells, we undertook a test of SCE formation in fibroblasts from 5 different FA complementation groups. In order to remove cell line variability, FA cells were tested in comparison to corrected controls generated using a complementing retroviral construct for each complementation group (except FA-D1). The FANCC and FANCG cells were primary cells, and the others were transformed lines. Although FA cells, like normal cells, show increased SCE formation with MMC treatment, no increase in SCE formation was noted in the FA cells compared to the corrected cells (fig. 8A, B). In addition, depletion of FANCA in BS cells did not increase SCE formation (fig. 2B). Thus human FA cells differ from BS cells in that they do not show increased SCEs relative to corrected cells. This outcome is in agreement with a report for human cells, but not in agreement with findings in immortalized chicken cells [Hirano et al., 2005; Godthelp et al., 2006].
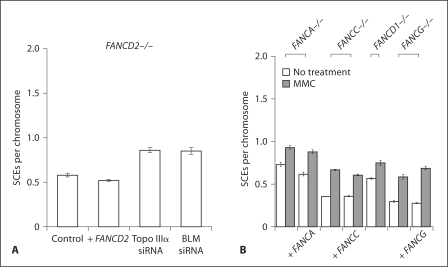
Fig. 8.
SCE formation in FA cells. FANCD2 (PD20) (A), FANCA (GM6914), FANCC (PD438F), FANCD1 (VU423), and FANCG (PD829) (B) were treated or not with MMC (20 ng/ml) and analyzed for SCE formation. Correction by complementing retroviral constructs is indicated, as is depletion by siRNA. Error bars represent standard error of the mean.
Depletion of BLM or Topo IIIα in FA-D2 cells produced an increase in SCEs, as it does in normal cells (fig. 8A). Therefore FA cells respond like normal cells to loss of BLM or Topo IIIα with an increase of SCE formation. These observations include the FANCD1 cell line, defective in BRCA2, as reported by others [Godthelp et al., 2006]. Therefore, the HR function of BRCA2 does not appear to be required for SCE formation (fig. 8B).
Fancd2–/–/Blmm3/m3 Mice Demonstrate Epistasis for FA and BS
The above results indicate that BLM is epistatic with the FA pathway for response to ICLs in human fibroblasts as measured by radial formation and cell survival. It might then be expected that the addition of a defect in BLM to an animal model for FA would not add detectable defects in the ICL response to the ones already noted. To test this, the hypomorphic mutant Blmm3/m3 allele [Luo et al., 2000] was combined with a Fancd2–/– genotype [Houghtaling et al., 2003] in a mixed background, since a complete defect in the murine BLM homolog is lethal [Chester et al., 1998]. By crossing heterozygotes, homozygous deficient mice were obtained at the predicted frequency of 1 in 16. The average weight of the double homozygous mutant mice was similar to that of either of the single mutants, indicating normal in utero growth. Primary fibroblast cells were established from the Blmm3/m3, Fancd2–/– mice and tested for their response to MMC. As observed in the BLM or Topo IIIα-depleted normal human cells or BS fibroblasts, murine fibroblasts lacking the BLM protein exhibited chromosomal instability in the form of increased radials. Furthermore, cells from the double mutant mice did not display increased radial formation above that of the Fancd2−/− mice (cells with one or more radial were 90% at 20 ng/ml MMC for Fancd2–/–/ Blmm3/+ versus 78% for Fancd2–/–/Blmm3/m3), supporting the epistatic relationship between BS and the FA pathway seen in human cells. In contrast, mouse cells defective for Blm alone, or both Blm and Fancd2 showed an increase in SCEs, while the cells defective in Fancd2 did not; that is, the fancd2 mutants did not show elevated SCE formation unless the blm hypomorphic mutation was added (table 1). These results support the conclusion that the FA pathway and Bloom pathway act epistatically in ICL repair, but that the FA path does not act in SCE regulation.
Table 1.
SCE analysis in mouse cells deficient for Blm and Fancd2. Errors represent standard errors of the mean, p values were calculated by an unpaired two-tailed t test.
| Mouse fibroblasts | Treatment | SCEs/chromosome | p value |
|---|---|---|---|
| Blm−/+ Fancd2−/+ | None | 0.64 + 0.05 | |
| Blm−/+ Fancd2−/− | None | 0.72 + 0.05 | 0.34 |
| Blm−/− Fancd2−/+ | None | 1.08 + 0.07 | <0.0001 |
| Blm−/− Fancd2−/− | None | 1.30 + 0.09 | <0.0001 |
Discussion
The results reported indicate that siRNA depletion can be used to demonstrate in human cells that the interaction of BLM and Topo IIIα is a functional one and that loss of either results in similar manifestations of genome instability following ICLs. BLM acts in several pathways for genome stability, including maintenance functions in response to breaks that arise during DNA replication and in response to DNA damage [Hickson, 2003]. However, the actions of BLM and Topo IIIα in response to ICLs are apparently limited to the FA pathway. In support of this, depletion of BLM or Topo IIIα in FA cells, or depletion of FANCA in BS cells, does not increase radial formation or decrease survival following MMC treatment. It is also known that inhibition of topoisomerase I does not increase the sensitivity of FA cells to MMC damage [Saito et al., 1994], and nor does depletion of Topo IIα, as shown here. Thus BLM and Topo IIIα appear to act specifically only with FANC proteins for ICL repair, defining BLM and Topo IIIα as epistatic to FA. In agreement with this proposal, fibroblasts from the mouse Blm/Fancd2 double mutant appear no more sensitive to MMC than the Fancd2 single mutant. These observations define a BLM-Topo IIIα-FA pathway for ICL response.
The picture is distinctly different for SCEs, since human FA cells do not show an increase in comparison to corrected cells, in agreement with observations made by others [Godthelp et al., 2006], but in contrast to a report on chicken cells [Hirano et al., 2005]. Our findings indicate that the function of BLM in control of SCEs is separate from its action in ICL repair. We chose to use human cells as a test system to avoid irregularities possibly arising from the very high HR frequency in the chicken cell system, where targeted HR rates are elevated as much as 10,000-fold [Winding and Berchtold, 2001]. Interestingly, SCE formation does not require BRCA2 function. This is surprising, given the supposed role of BRCA2 in HR [Shivji and Venkitaraman, 2004] and the apparent dependence of SCE formation on HR [Sonoda et al., 1999]. This would suggest that SCEs arising from BLM deficiency arise via a BRCA2-independent branch of the HR system.
BLM has been shown to be phosphorylated by ATM or ATR [Davies et al., 2004], but phosphorylation, which is dependent on a functional FANC core [Pichierri et al., 2004], is not required for its action in SCE regulation; rather it is required for other functions such as recovery from S-phase arrest [Davies et al., 2004]. Therefore, BLM, and presumably its partner, Topo IIIα, have segregated functions in ICL repair, versus regulation of SCE formation. In summary, we have provided evidence for the existence of a functionally important ICL response network comprising the BLM, Topo IIIα and FA proteins.
Acknowledgements
We thank D. Bruun for helpful conversations. This work was supported by National Institutes of Health grants PO1 HL048546, CA52121, and GM08796. P.S.N and I.D.H. are supported by Cancer Research, UK.
References
- Bennett RJ, Noirot-Gros MF, Wang JC. Interaction between yeast Sgs1 helicase and DNA topoisomerase III. J Biol Chem. 2000;275:26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat Genet. 2005;37:953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- Bruun D, Folias A, Akkari Y, Cox Y, Olson S, Moses R. siRNA depletion of BRCA1, but not BRCA2, causes increased genome instability in Fanconi anemia cells. DNA Repair (Amst) 2003;2:1007–1013. doi: 10.1016/s1568-7864(03)00112-5. [DOI] [PubMed] [Google Scholar]
- Bussen W, Raynard S, Busygina V, Singh AK, Sung P. Holliday junction processing activity of the BLM-Topo IIIalpha-BLAP75 complex. J Biol Chem. 2007;282:31484–31492. doi: 10.1074/jbc.M706116200. [DOI] [PubMed] [Google Scholar]
- Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci USA. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraverty RK, Kearsey JM, Oakley TJ, Grenon M, de La Torre Ruiz MA, et al. Topoisomerase III acts upstream of Rad53p in the S-phase DNA damage checkpoint. Mol Cell Biol. 2001;21:7150–7162. doi: 10.1128/MCB.21.21.7150-7162.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, et al. Rmi1/Nce4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester N, Kuo F, Kozak C, O'Hara CD, Leder P. Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Bloom's syndrome gene. Genes Dev. 1998;12:3382–3393. doi: 10.1101/gad.12.21.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester N, Babbe H, Pinkas J, Manninig C, Leder P. Mutation of the murine Bloom's syndrome gene produces global genome destabilization. Mol Cell Biol. 2006;17:6713–6726. doi: 10.1128/MCB.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol. 2007;14:677–679. doi: 10.1038/nsmb1267. [DOI] [PubMed] [Google Scholar]
- Folias A, Matkovic M, Bruun D, Reid S, Hejna J, et al. BRCA1 interacts directly with the Fanconi anemia protein FANCA. Hum Mol Genet. 2002;11:2591–2597. doi: 10.1093/hmg/11.21.2591. [DOI] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Gaymes TJ, North PS, Brady N, Hickson ID, Mufti GJ, Rassool FV. Increased error-prone non homologous DNA end-joining – a proposed mechanism of chromosomal instability in Bloom's syndrome. Oncogene. 2002;21:2525–2533. doi: 10.1038/sj.onc.1205331. [DOI] [PubMed] [Google Scholar]
- German J. Bloom's syndrome. Dermatol Clin. 1995;13:7–18. [PubMed] [Google Scholar]
- Godthelp BC, van Buul PP, Jaspers NG, Elghalbzouri-Maghrani E, van Duijn-Goedhart A, et al. Cellular characterization of cells from the Fanconi anemia complementation group, FA-D1/BRCA2. Mutat Res. 2006;601:191–201. doi: 10.1016/j.mrfmmm.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci. 2007;64:2306–2322. doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson ID. RecQ helicases: Caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- Hirano S, Yamamoto K, Ishiai M, Yamazoe M, Seki M, et al. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J. 2005;24:418–427. doi: 10.1038/sj.emboj.7600534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, et al. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17:2021–2035. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow JK, Wu L, Hickson ID. RecQ family helicases: Roles in cancer and aging. Curr Opin Genet Dev. 2000;10:32–38. doi: 10.1016/s0959-437x(99)00039-8. [DOI] [PubMed] [Google Scholar]
- Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005;37:934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- Li W, Wang JC. Mammalian DNA topoisomerase IIIalpha is essential in early embryogenesis. Proc Natl Acad Sci USA. 1998;95:1010–1013. doi: 10.1073/pnas.95.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, et al. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- McDaniel LD, Chester N, Watson M, Borowsky AD, Leder P, Schultz RA. Chromosome instability and tumor predisposition inversely correlate with BLM protein levels. DNA Repair (Amst) 2003;2:1387–1404. doi: 10.1016/j.dnarep.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, et al. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol. 2003;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses RE. DNA damage processing defects and disease. Annu Rev Genomics Hum Genet. 2001;2:41–68. doi: 10.1146/annurev.genom.2.1.41. [DOI] [PubMed] [Google Scholar]
- Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuki M, Seki M, Inoue E, Abe T, Narita Y, et al. Analyses of functional interaction between RECQL1, RECQL5, and BLM which physically interact with DNA topoisomerase III alpha. Biochim Biophys Acta. 2008;1782:75–81. doi: 10.1016/j.bbadis.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Pichierri P, Franchitto A, Rosselli F. BLM and the FANC proteins collaborate in a common pathway in response to stalled replication forks. Embo J. 2004;23:3154–3163. doi: 10.1038/sj.emboj.7600277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- Saito H, Grompe M, Neeley TL, Jakobs PM, Moses RE. Fanconi anemia cells have a normal gene structure for topoisomerase I. Hum Genet. 1994;93:583–586. doi: 10.1007/BF00202828. [DOI] [PubMed] [Google Scholar]
- Schmidt KH, Kolodner RD. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol Cell Biol. 2004;24:3213–3226. doi: 10.1128/MCB.24.8.3213-3226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TM, Anschutz F, Knopp A. Spontaneous chromosome aberrations in familial panmyelopathy. Humangenetik. 1964;1:194–196. doi: 10.1007/BF00389636. [DOI] [PubMed] [Google Scholar]
- Shivji MK, Venkitaraman AR. DNA recombination, chromosomal stability and carcinogenesis: Insights into the role of BRCA2. DNA Repair (Amst) 2004;3:835–843. doi: 10.1016/j.dnarep.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, et al. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, et al. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell. 2001;7:241–248. doi: 10.1016/s1097-2765(01)00172-1. [DOI] [PubMed] [Google Scholar]
- Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, et al. Mutation in BRCA2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Winding P, Berchtold MW. The chicken B cell line DT40: A novel tool for gene disruption experiments. J Immunol Methods. 2001;249:1–16. doi: 10.1016/s0022-1759(00)00333-1. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu Rev Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- Wu L, Bachrati CZ, Ou J, Xu C, Yin J, et al. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci USA. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Guo R, Sobeck A, Bachrati CZ, Yang J, et al. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008;22:2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]