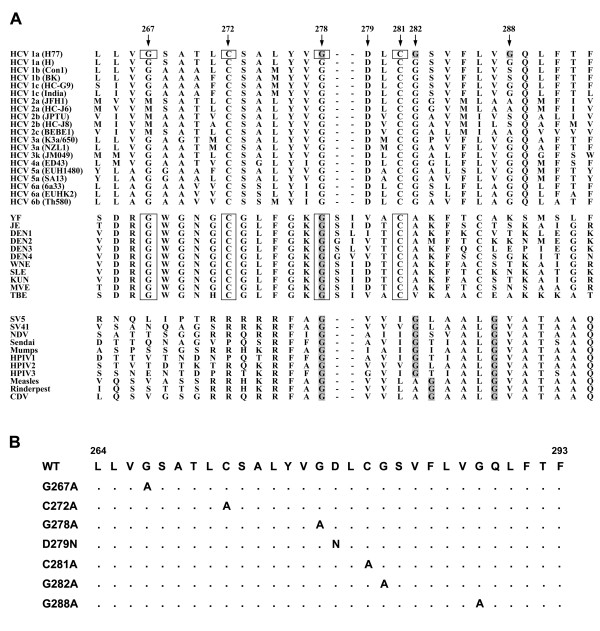
Figure 1.
Analysis of the HCV E1 putative fusion domain. (A) Alignment of the E1 fusion peptide-like motif of HCV with the predicted fusion peptide sequences of the E protein of flaviviruses and fusion protein of paramyxoviruses. The E1 putative fusion peptide sequence derived from various HCV genotypes contains similarities to the predicted fusion peptide of flavivirus E glycoproteins which are boxed. The spacing of Gly residues, as indicated by shading in the HCV putative fusion peptide, is also similar to those within the fusion peptides of paramyxovirus F proteins. (B) Construction of E1 fusion peptide-like domain mutants. Construction of E1 mutants with substitutions of Ala or Asn for the residues as indicated in the fusion peptide-like domain was performed by a site-specific, oligonucleotide-directed PCR overlap extension method as described in "Materials and Methods". Dots in the sequence indicate that the amino acid in that position is identical to that of the WT sequence.