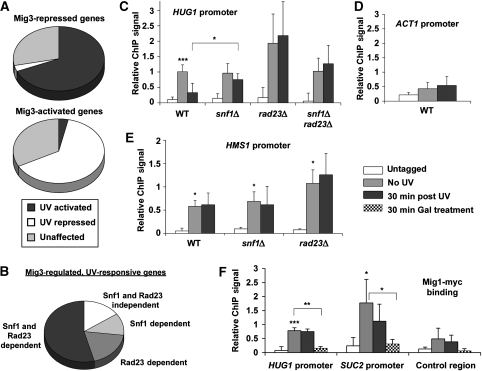
Figure 6.
Mig3 directly regulates UV-responsive, Snf1- and Rad23-regulated genes. (A) UV dependence of Mig3-regulated genes. Comparison of microarray data sets for UV-irradiated WT cells with the unirradiated mig3Δ data set shows the proportion of Mig3-regulated genes that were significantly affected by UV irradiation. (B) Snf1- and Rad23-dependence of the UV response of Mig3-regulated genes. Comparison of the UV response in snf1Δ, rad23Δ, and snf1Δ rad23Δ cells with WT for Mig3-regulated, UV-responsive genes indicates that most were Snf1- or Rad23-dependent. (C–E) Mig3 binding to the promoters of Mig3-regulated genes. ChIP and RT–PCR were performed for Myc-tagged Mig3 using primers specific for the HUG1 promoter (C), ACT1 promoter (D), and HMS1 promoter (E). Relative ChIP signal was obtained by subtracting mock IP samples and normalizing to the total input chromatin for each sample. ChIP signal was calculated for cells containing untagged Mig3 as a negative control and cells harbouring Myc-tagged Mig3 before irradiation and 30 min after irradiation. Values are the average of three independent experiments and error bars represent standard deviation. Statistical significance was measured using a two-tailed Student's t-test with paired variables (***P<0.01, **P<0.03, *P<0.05; compared to untagged unless otherwise noted). Although there was significant variability associated with the triplicate ChIP results obtained using rad23Δ cells, the failure of Mig3 to vacate the HUG1 promoter in rad23Δ cells after irradiation (panel C) was highly reproducible (average ratio of ChIP signal for irradiated/unirradiated cells was 1.13±0.16). Thus, the error bars reflect the day-to-day variability in the Mig3 occupancy versus untagged control rather than variation in the Mig3 occupancy when comparing samples with and without UV treatment prepared in parallel. (F) Mig1 binding to Mig3- and Mig1-regulated promoters. ChIP and RT–PCR were carried out for Myc-tagged Mig1 using primers specific for the HUG1 promoter, SUC2 promoter, and an intergenic region on chromosome V. ChIP signal was obtained as in panels (C–E).