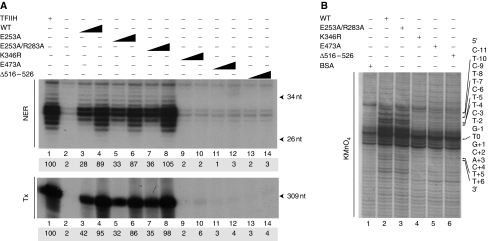
Figure 4.
Mutations in R-E-D and ThM motifs impair the ATPase activity of XPB. (A) A measure of 25 and 75 ng of TFIIH(WT), TFIIH/XPB(E253A), XPB(E253A/R283A), XPB(K346R), XPB(E473A) or XPB(Δ516–526) was tested in a dual incision assay (NER, upper panel) or in a reconstituted transcription assay (Tx, lower panel) as described (Coin et al, 2004). Lane 1 contains highly purified Hela TFIIH (Giglia-Mari et al, 2004). Lane 2 contains all the factors except TFIIH. The sizes of the incision or transcription products are indicated. The transcription and repair signals were quantified using Genetool (Syngene). (B) A measure of 100 ng of the various TFIIH complexes were tested in a KMnO4 footprint assay (see Figure 1B). Lane 1; Pt-DNA with BSA only. Residues are numbered with the central thymine of the crosslinked GTG sequence designated T0. Arrows indicate KMnO4 sensitive sites. Adducted strand residues to the 3′ and 5′ of T0 are denoted by positive and negative integers (+N, –N).