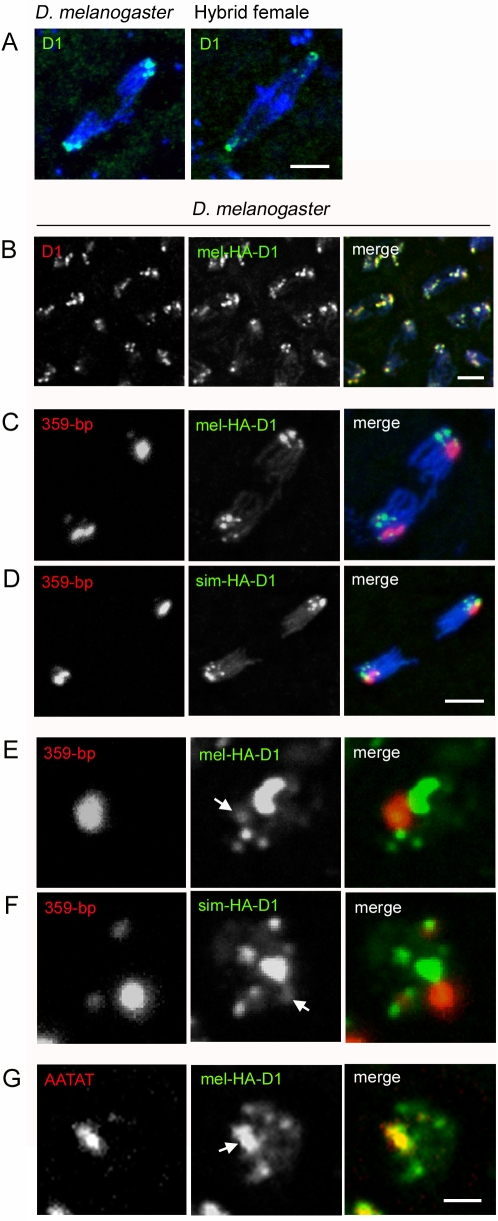
Figure 4. D1 localizes to AATAT satellite DNA but not the 359-bp satellite block during early embryogenesis.
(A) Left panel, an anaphase spindle in a D. melanogaster embryo. Anti-D1 highlights endogenous D1, which localizes to pericentromeric regions near the spindle poles. Right panel, an anaphase spindle in a hybrid female embryo. Endogenous D1 exhibits a pattern similar to that found in D. melanogaster. DNA is blue in both panels. (B) Anaphase spindles from a D. melanogaster embryo with HA-tagged D. melanogaster D1 (UAS-mel-HA-D1 driven by P{matα4-GAL-VP16}V37). Anti-D1 recognizes both endogenous and HA-D1. The two signals overlap completely, showing that the HA-D1 protein localizes identically to endogenous D1. (C) Anaphase spindle from the same genotype in (B), showing mel-HA-D1 and 359-bp DNA. (D) Anaphase spindle from a D. melanogaster embryo with HA-tagged D. simulans D1 (UAS-sim-HA-D1 driven by P{matα4-GAL-VP16}V37). Neither of the D1 orthologs, (C) and (D), co-localizes with the 359-bp satellite block. (E) Interphase nucleus from the same genotype in (C). (F) Interphase nucleus from the same genotype in (D). Both D1 orthologs show only a slight co-localization with the 359-bp satellite block during interphase. White arrows in (E) and (F) indicate this minimal co-localization. (G) Interphase nucleus from the same genotype in (C). The mel-HA-D1 protein co-localizes strongly with AATAT satellite DNA (indicated by white arrow). Scale bar is 4 µm in (A), 5 µm in (B), 4 µm in (C, D) and 3 µm in (E–G).