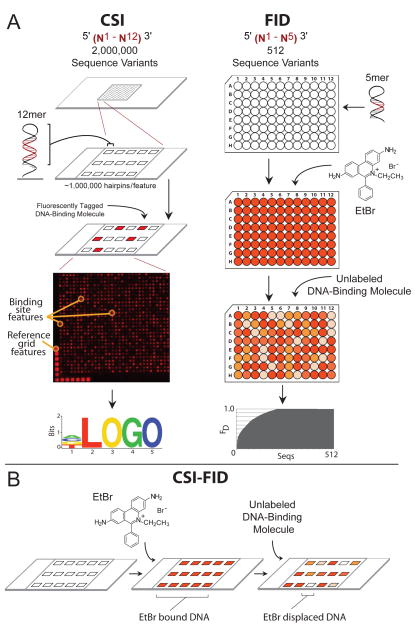
Figure 1. CSI-FID combines CSI and FID for label free detection on highly complex DNA libraries.
A. The left panel illustrates the CSI array technology. First a high density oligonucleotide array (up to two million sequence variants) is synthesized using maskless array synthesis. The oligos are hairpinned to form dsDNA, and a fluorescently tagged DNA-binding molecule is applied to the array. A readout of fluorescent intensities is then used to generate a sequence motif of the highest bound DNA sequences. The right panel shows the procedure for the FID assay. First a library of hairpinned DNA oligos are individually arrayed in 96 well plates, with one DNA hairpin sequence per well. EtBr is then added to each well and the fluorescence is measured. Next an unlabeled DNA molecule of interest is applied to the plate and the diplacement of EtBr from the DNA is determined. The sequences with the highest EtBr displacement bind the molecule of interest with the highest affinity and these sites are used to generate a consensus motif for the molecule. B. Depiction of FID being adapted for use on CSI arrays to generate the CSI-FID assay. First EtBr is incubated with the CSI array, then an unlabeled molecule of interest is applied to the array. The ratios of the EtBr intensity for the displaced versus undisplaced values for each DNA feature provides a measurement of the affinity of the small molecule for each DNA sequence. This technology measures the sequence specific binding for unlabelled DNA-binding molecules on CSI microarrays bearing highly complex DNA libraries.