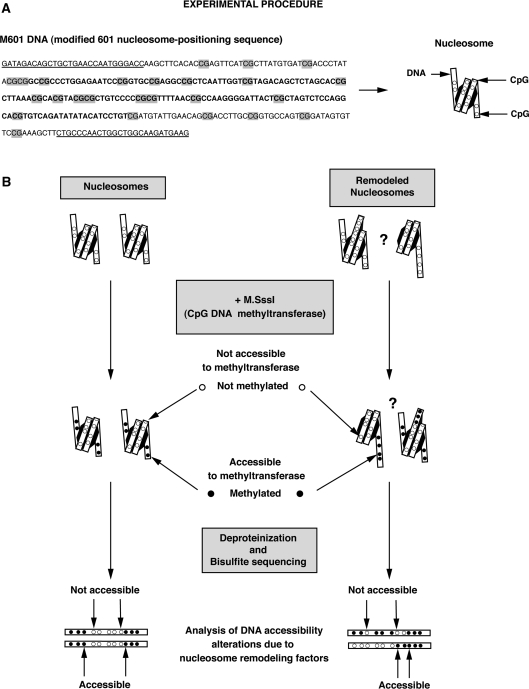
Figure 1.
(A) M601 DNA sequence used in the remodeling assays. GRP78-gene primer sequences flanking the DNA construct are underlined and CpG dinucleotides are highlighted in grey. The residues in bold denote the approximate position of the protection caused by the histone octamer on the nucleosome substrate. (B) Schematic representation of the procedure used to perform single molecule analyses of the remodeled products. Nucleosomes were assembled using the M601 DNA. After incubation with (or without) nucleosome remodeling factor, remodeled nucleosomes were methylated with the M.SssI CpG methyltransferase to create a footprint of DNA accessibility (the DNA methyltransferase only methylates cytosine residues that are not bound to the histones). After native electrophoresis, excision and elution from the gel, nucleosomes were deproteinized and the nucleosomal DNA molecules subjected to conventional bisulfite treatment to reveal their methylation pattern. Changes in these patterns were compared to the input nucleosome to assess alterations in histone–DNA contacts resulting from the action of the remodeling factors.