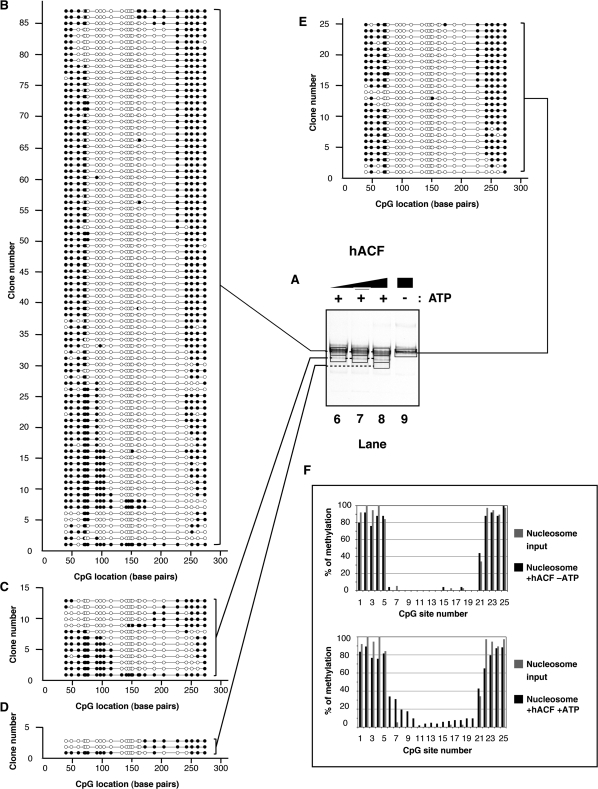
Figure 4.
(A) Native electrophoresis of hACF remodeled products. Nucleosomes (∼100 nM) were incubated with increasing concentrations of hACF complex (lane 6: 8 nM; lane 7: 24 nM; lanes 8 and 9: 72 nM) in the presence (lanes 6–8) or absence (lane 9) of ATP (1 mM) as indicated on top. Reactions were handled identically and in parallel to samples in Figure 3. (B–E) Schematic representation of individual DNA molecules remodeled by hACF. Bisulfite-converted DNAs from gel slices (black frames, lanes 6–9) were amplified by PCR, cloned and sequenced. Individual DNA clones are represented as described in Figure 3. (F) Frequency of methylation at a given CpG site. Upper panel: the frequency of methylation was determined by averaging methylation for all the DNA molecules showed in panel E (reaction without ATP). Lower panel: the frequency of methylation was determined by averaging methylation for all the DNA molecules showed in panels B–D (reactions with ATP). In both the upper and lower panels, frequencies obtained from the nucleosome substrate in the absence of remodeler (Figure 2E) are shown in grey for comparison.