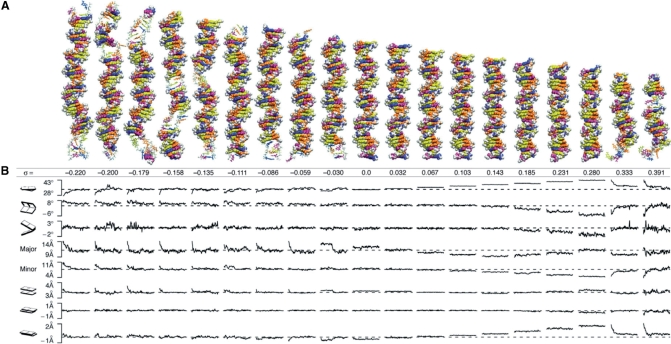
Figure 1.
Structures and time courses of twisted DNA. (A) Atomic structures of DNA as a function of underwinding or overwinding, σ, that have minimum RMSD from the average simulation structure. Regions of structural failure are rendered with sticks and regions of Watson–Crick base pairing are rendered with van der Waals spheres. The bases are colored: adenine, blue; thymine, purple; guanine, yellow; and cytosine, orange. Backbone atoms are in white. The σ = 0.0 (Lk0) structure is a control showing that the simulation conditions, including the boundary, had no remarkable effect on the B-DNA structure. As an additional test of the boundary conditions, we translated the σ = −0.059 and σ = 0.391 systems through the periodic boundaries and were able to reproduce base flipping and P-DNA events, respectively. (B) Structural parameters (twist, roll, tilt, major groove, minor groove, rise, shift and slide) averaged over the sequence from snapshots taken every 100 ps over the total simulation time of 10 ns. The dashed lines are the values of a relaxed helix with the same sequence, as predicted by analysis of PDB structures (22).