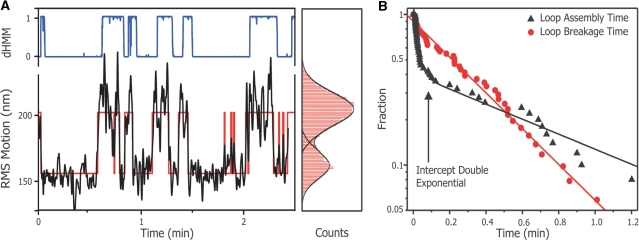
Figure 5.
Obtaining looping kinetics with thresholding analysis. (A) Shown are a RMS time trace (left panel, black line) and its corresponding histogram (right panel, red bars) of a single two-site DNA tether in the presence of 0.1 nM D79A in Mg2+ buffer. The histogram is fitted with a double Gaussian (right panel, black lines) and the intersection of the double Gauss is used as a thresholding value. The thresholding results in a binary trace (left panel, red line), from which the dwell times are extracted. Also shown is the probability for each data point to belong to the unlooped distribution (blue line between) as obtained by the diffusive hidden Markov analysis. (B) A CDF plot of the obtained dwell times. The dwell times displayed are from a single tether in the presence of 0.1 nM D79A in Mg2+ buffer, and contain at least 50 data points. The red circles show the distribution of the loop lifetime and are fitted with a single exponential (red line) to obtain the loop breakage rate. The loop assembly times are shown by black triangles and fitted with a bi-exponential decay. The intersection of the two exponentials (arrow) from the bi-exponential fit can be used to estimate the protein dissociation rate of the D79A protein from the DNA.