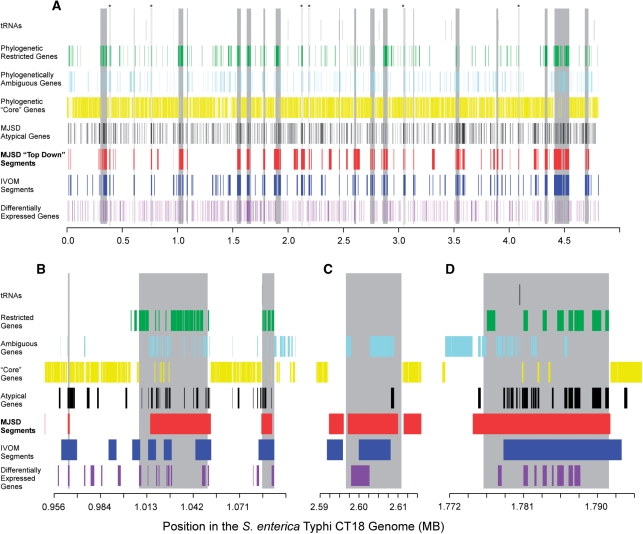
Figure 6.
Predictions made by the top–down and gene-based MJSD, IVOM, and phylogenetic methods. Known and novel islands are shown as vertical gray bars. Genes restricted to the Salmonella genome and those found in related genomes (‘core’ genes) were detected by BLAST similarities (see ‘Materials and Methods’ section). Differentially expressed genes are those upregulated during pathogenesis (26). (A) The entire genome of S. enterica Typhi CT18 showing previously described islands (Supplementary Table 3). (B) A region of the genome that demonstrates the ability of top–down MJSD to accurately capture island boundaries and identify novel regions of varying sizes. The two known islands and a likely non-functional islet (the small region near 965 kb) are accurately detected. (C) The CS54 island's boundaries are best defined using top–down MJSD. (D) An island composed of an integrase gene and known virulence genes is delineated by both the MJSD and IVOM methods; borders are approximated.