Figure 1.
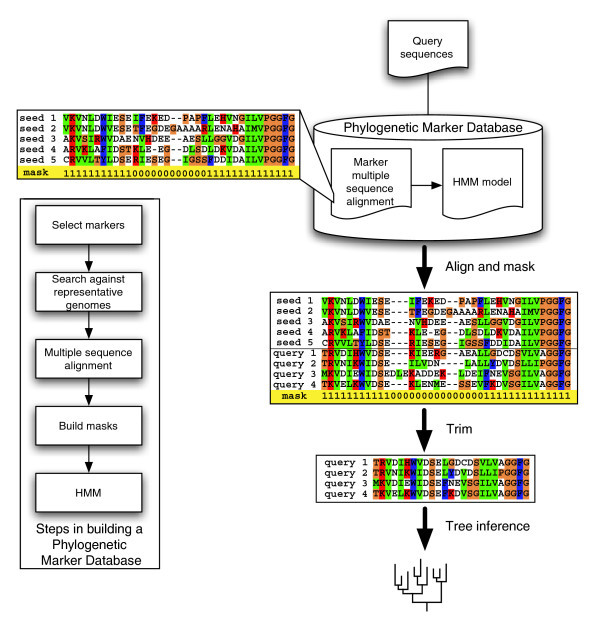
A flowchart illustrating the major components of AMPHORA. The marker protein sequences from representative genomes are retrieved, aligned, and masked. Profile hidden Markov models (HMMs) are then built from those 'seed' alignments. New sequences of interest are rapidly and accurately aligned to the trusted seed alignments through HMMs. Predefined masks embedded within the 'seed' alignment are then applied to trim off regions of ambiguity before phylogenetic inference. Alignment columns marked with '1' or '0' were included or excluded, respectively, during further phylogenetic analysis.
