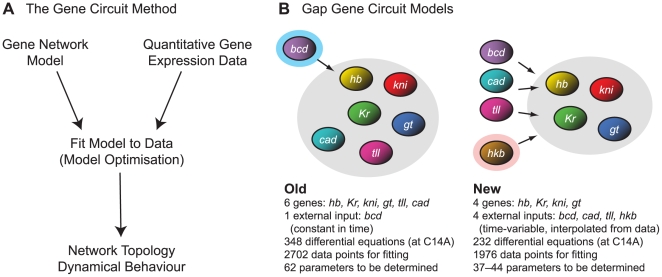
Figure 1. The gene circuit method: old vs. new models.
(A) Reverse engineering gene regulatory networks using the gene circuit method: A mathematical (dynamical) model of the network is fit to quantitative, spatial gene expression data using combined global and local non-linear optimisation approaches. The resulting gene circuits, consisting of specific estimated sets of parameter values, define regulatory interactions among genes within the network (its regulatory topology). This topology is not defined a priori, but is extracted from the quantitative expression data by the fitting procedure. The resulting dynamical behaviour of the system can be analysed using graphical or numerical methods. (B) Previous gap gene circuit models used concentrations of the protein products of gap genes hb, Kr, gt, kni, tll, and of the maternal co-ordinate gene cad as state variables (grey shaded background), while the maternal protein gradient encoded by bcd was implemented as an external input which did not vary over time (6-gene models, left; time-constant Bcd indicated by blue shaded background). Current gap gene circuit models only include the trunk gap genes hb, Kr, gt and kni as state variables (grey shaded background), implementing bcd, cad, tll and hkb as time-variable external inputs since they are not regulated by gap genes themselves (4-gene models, right). hkb (highlighted), which is the focus of this study, has not been considered in previous models. See main text for details.