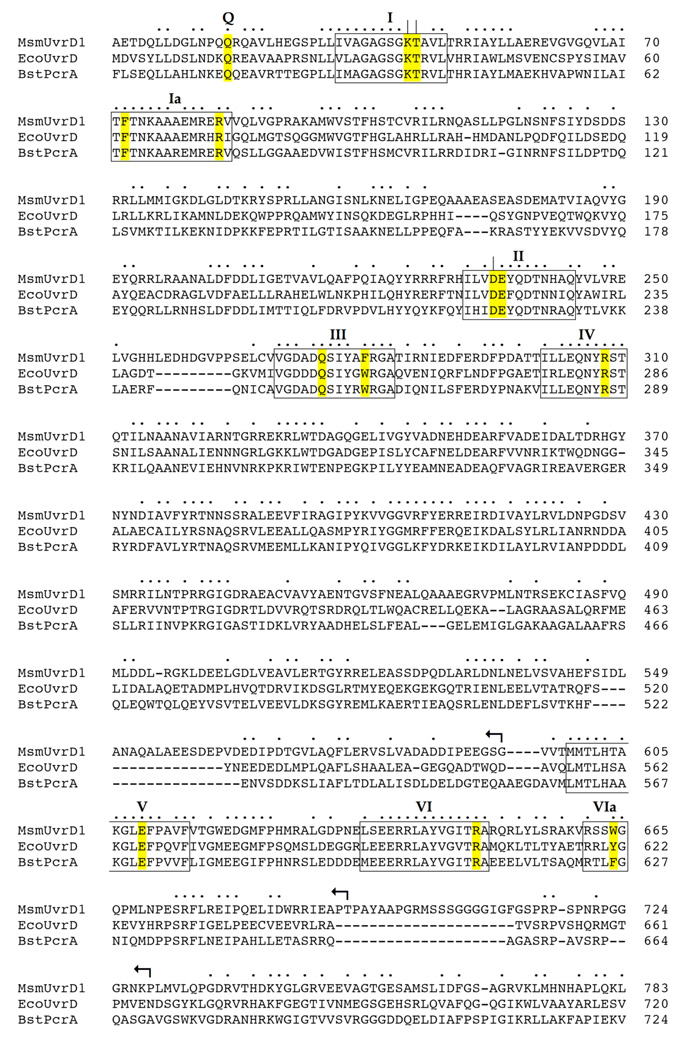
Fig. 1. Structural similarity between UvrD1, UvrD and PcrA.
The amino acid sequence of M. smegmatis (Msm) UvrD1 (accession YP_889771) is aligned to the sequences of E. coli (Eco) UvrD (accession NP_418258) and B. stearothermophilus (Bst) PcrA (accession P56255). Positions of side chain identity/similarity in all three proteins are indicated by •. Gaps in the alignment are denoted by –. The ATPase/helicase motifs are denoted in boxes and named according to Lee and Yang (4). The conserved residues comprising the ATPase active site and DNA-binding interfaces of UvrD/PcrA that were subjected to alanine scanning in UvrD1 are highlighted in yellow. Essential constituents of UvrD1 ATPase motifs I (Lys45, Thr46) and II (Asp235) that were identified previously are indicated by |. The targeted UvrD1 residues, and their equivalents in UvrD and PcrA, are listed in Table I. The C-terminal truncations of UvrD1 are denoted by ↰.