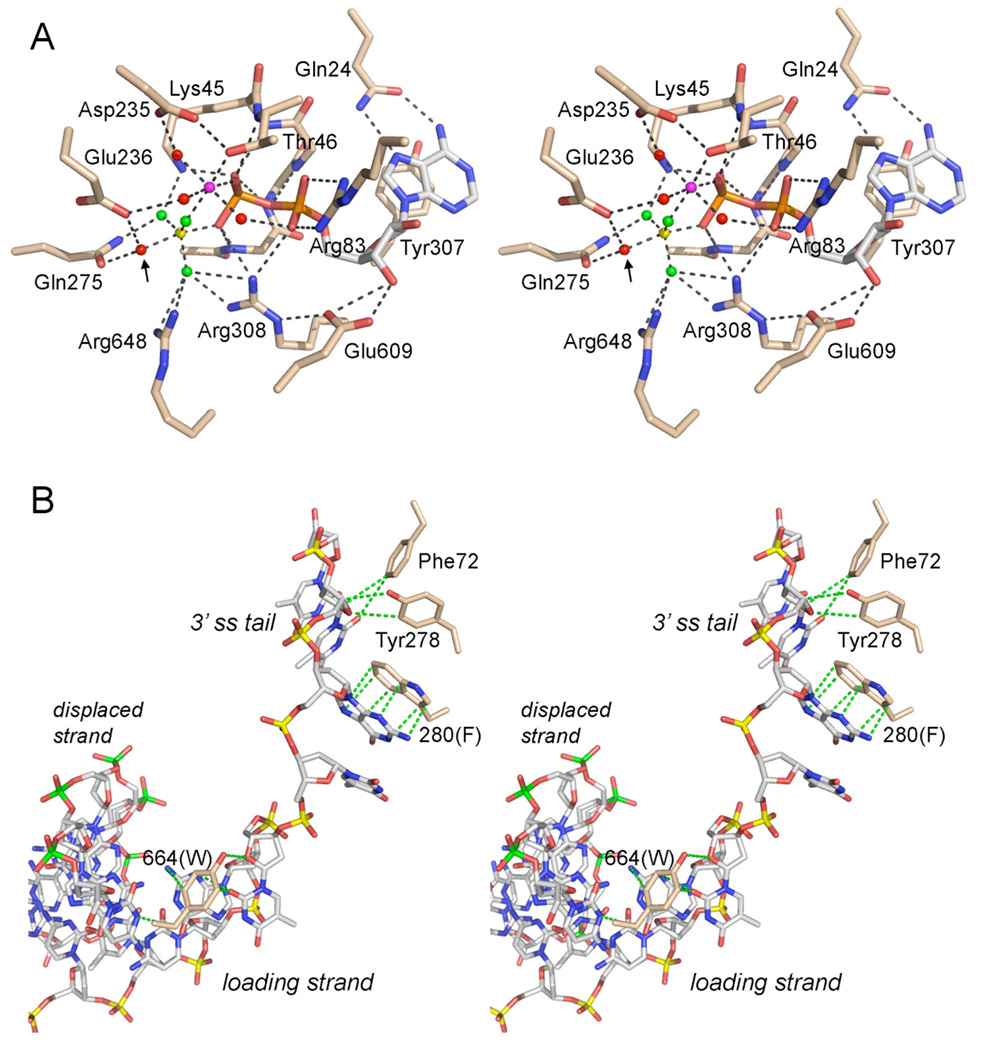
Fig. 2. ATPase active site and DNA contacts.
(A) Stereo view of the phosphohydrolase active site of DNA-bound E. coli UvrD (PDB 2IS6) as a transition state mimetic in complex with Mg2+ (magenta sphere), ADP (stick model with gray carbons), and a trigonal planar MgF3 (depicted with the magnesium as a yellow sphere and the fluorines as green spheres). Waters are rendered as red spheres. The putative water nucleophile located apical to the leaving β-phosphate oxygen is indicated by an arrow. The atomic contacts of the conserved active site residues (stick models with beige carbons) are denoted by dashes lines. The residue numbers refer to the equivalent side chains in UvrD1 that were subjected to alanine scanning. (B) Stereo view of DNA contacts in the crystal structure of E. coli UvrD bound to a 3’ tailed duplex DNA (from PDB 2IS6). The 3’ tailed strand (the loading strand for initial helicase binding, on which the enzyme translocates 3’ to 5’) is depicted as a stick model with phosphorus atoms colored yellow. The complementary strand that forms the duplex segment (the displaced strand unwound by helicase translocation) is depicted with phosphorus atoms in green. Conserved aromatic residues that make base stacking and van der Waals contacts with the DNA (green dashed lines) are shown, with residue numbering referring to the equivalents in UvrD1.