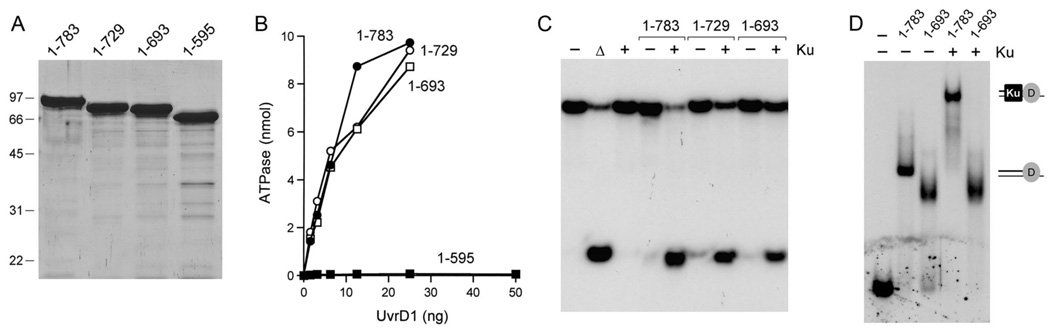
Fig. 6. Effects of C-terminal deletions.
(A) Aliquots (5 µg) of full-length UvrD1-(1–783) and the C-terminal truncation mutants UvrD1-(1–729), UvrD1-(1–693) and UvrD1-(1–595) were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The sizes (kDa) and positions of marker proteins are indicated on the left. (B) ATPase assays were performed as described in Methods. 32Pi release from 1 mM [γ32P]ATP is plotted as a function of input UvrD1 for each enzyme assayed. (C) Complete helicase reaction mixtures contained 1 mM ATP, 50 nM 32P-labeled tailed duplex DNA substrate, 75 ng Ku (where indicated by +) and 100 ng UvrD1 protein as specified. The products were analyzed by native PAGE and visualized by autoradiography. A reaction lacking protein that was heat-denatured prior to PAGE is shown in lane Δ. (D) DNA binding reaction mixtures contained 0.5 pmol 32P-labeled 3’-tailed DNA, 100 ng UvrD1-(1–783) or UvrD1-(1–693), and 75 ng Ku (where indicated by +). The free DNA and protein-DNA complexes (depicted on the right) were resolved by native PAGE and visualized by autoradiography.