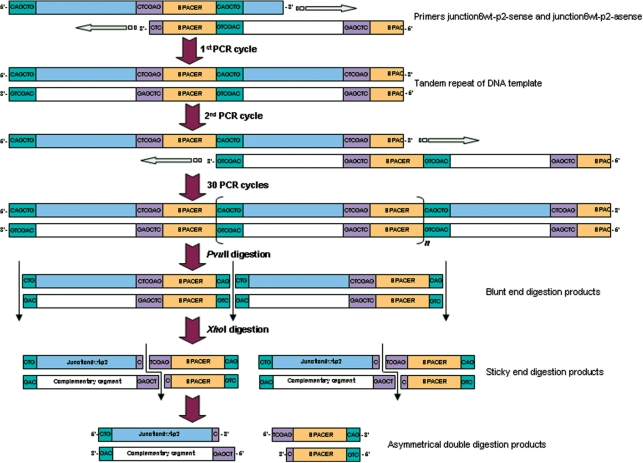
Figure 1.
Schematic of the proposed primer-pair amplification by self-primed PCR, here shown for the 20-nt junction 6 wt-p2 ssDNA sequence (see below). The two designed primers hybridize to form an amplifiable overlap that is filled-in by Pfu DNA polymerase in the first PCR cycle. The formed tandem repeat of dsDNA will be elongated in the next stages of cycling by the possibility to shift annealing position of the tandem repeated dsDNA. After 30 cycles, the long DNA is digested with a blunt-end-generating endonuclease followed by digestion with a sticky-end-generating endonuclease. The asymmetry in the digested DNA molecules allows for separation of the four individual strands on denaturing PAGE.