Abstract
Damages in the DNA template inhibit the progression of replication, which may cause single-stranded gaps. Such situations can be tolerated by translesion DNA synthesis (TLS), or by homology-dependent repair (HDR), which is based on transfer or copying of the missing information from the replicated sister chromatid. Whereas it is well established that TLS plays an important role in DNA damage tolerance in mammalian cells, it is unknown whether HDR operates in this process. Using a newly developed plasmid-based assay that distinguishes between the three mechanisms of DNA damage tolerance, we found that mammalian cells can efficiently utilize HDR to repair DNA gaps opposite an abasic site or benzo[a]pyrene adduct. The majority of these events occurred by a physical strand transfer (homologous recombination repair; HRR), rather than a template switch mechanism. Furthermore, cells deficient in either the human RAD51 recombination protein or NBS1, but not Rad18, exhibited decreased gap repair through HDR, indicating a role for these proteins in DNA damage tolerance. To our knowledge, this is the first direct evidence of gap-lesion repair via HDR in mammalian cells, providing further molecular insight into the potential activity of HDR in overcoming replication obstacles and maintaining genome stability.
INTRODUCTION
During DNA replication, damaged template nucleotides hinder the progression of the replication machinery. Stalled replication forks may either be resolved directly, by the recruitment of specialized proteins, or replication could be re-initialized further-on resulting in the formation of a single stranded gap. In this context, damaged nucleotides cannot be repaired through excision, and continuing replication necessitates the enlistment of DNA damage tolerance mechanisms, which restore the double stranded structure of the DNA (1–4).
The tolerance of DNA damage may occur in one of two possible mechanisms. These are translesion DNA synthesis (TLS) and homology-dependent repair (HDR) [We use the term HDR for all repair mechanisms that rely on a homologous donor; this includes strand transfer from the donor (commonly termed homologous recombination repair, HRR), and template switch mechanisms, in which the information is copied from the donor] (1). In TLS, specialized DNA polymerases insert a nucleotide across from the damage. However, the miscoding nature of damaged templates renders such a process inherently error prone (5–7). In HDR, the missing information is transferred from a homologous molecule, be it the homologous chromosome or the sister chromatid. While in bacteria and yeast, homologous recombination (HR) in general appears to be prevalent, the situation in somatic mammalian cells is less clear. Despite reports on low efficiencies of homology-directed sequence integration into the genome (8–10) and spontaneous recombination events (11–14), actual recombination efficiencies are difficult to measure, and may in fact be higher than it appears from those studies. The underlying logic for low HR in mammalian cells relies on the possible deleterious consequences of illegitimate HR events. For instance, a recombination event between repetitive sequences on different chromosomes could cause gross chromosomal aberrations. However, this does not exclude the possibility of highly efficient HR mechanisms under certain conditions.
HDR has been established to play a role in DNA double-strand break (DSB) repair in mammalian cells (15–18). This role appears to be minor compared to the alternative repair mechanism, non-homologous end joining (NHEJ) (19–23). However, HDR is also induced by agents that neither form DSB nor induce NHEJ. These include alkylating agents, heavy metals, agents that form bulky adducts (e.g. benzo[a]pyrene; BP), cross linkers and UV light (17,24–26). This implies that a HDR may be utilized for the repair of DNA lesions other then DSB. Recent studies suggest that stalled replication forks are indeed substrates for HDR; however, this was assayed in the absence of a chemical DNA damage (17,26–28).
While in Escherichia coli and Saccharomyces cerevisiae the operation of HDR is well established (1), and estimated to be responsible for most DNA damage tolerance events (29–32), the existence of such a process in mammalian cells has not been yet proven. A mechanism utilizing HR for gap-lesion filling is certainly feasible in mammalian cells as all the necessary machinery is present. Homologs and paralogs of most yeast recombination proteins exist in mammalian cells (33,34). Furthermore, an activity of HDR is expected to be needed either during or immediately after replication, when a sister chromatid is in cohesion, thus minimizing the risk for an illegitimate event. In addition, in contrast to the alternative TLS mechanism, it does not bear an inherent mechanistic mutagenic property (although under certain conditions it can cause gross DNA changes such as rearrangements and deletions).
In this study, we set out to establish whether HDR can play a role in DNA damage tolerance in mammalian cells. In order to do so, we utilized a plasmid-based assay system as a model for post-replicative gap repair. We show that mammalian cells can efficiently utilize HDR to fill in gaps opposite lesions. The process occurs primarily via a strand transfer rather than template switch mechanism, and involves RAD51 and NBS1 but not Rad18.
MATERIALS AND METHODS
Cell culture
The human large cell lung cancer cell line H1299 (35) was grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. The human colon cancer cell line HCT116 (36,37) was cultured in McCoy's 5A medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. The SV40 transformed NBS1 fibroblasts and their complemented controls (37) and the Rad18+/+ and Rad18−/− MEF (38) were grown in DMEM media supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin.
Construction of DNA substrates
The gapped plasmids (GPs) were synthesized as previously described (39,40). Homologous linear DNA donors were synthesized by restriction of closed circular plasmids identical in sequence to filled GPs (pFGP) bearing either cmR or ampR instead of the kanR genes. The homologous donor DNA (hDNA) is the 1072 restriction product of the digestion of either plasmid with the DraI and HindIII enzymes. The homologous plasmids used as donors for GP-abasic (pFGP-C-amp and pFGP-T-amp) were ampR kanS derivatives of previous TLS experiments. The construction of these plasmids was conducted by digestion of pFGP21-C/T with XhoI and HindIII, deleting a 520-bp fragment from the Kan gene, and ligating the restricted plasmids to a 945-bp fragment carrying PCR termini with XhoI and HindIII sites, and the bla gene from plasmid pUC18 conferring ampR. The additional hDNA molecules were obtained from closed circular pFGP plasmids bearing cmR (pFGP-cm). In order to obtain these plasmids, cmR vector DNA (the BstXI–BsaI restriction product of pSKSL-cm) was ligated to an appropriate double-stranded insert DNA (achieved by annealing of two oligonucleiotides) in order to achieve the sequence homologous to the specific GP but with the additional recombination markers. For hDNA-BPG, the two oligonucleotides were: 5′-ACCGCAACGAAGTGATTCGGCATCCGTCCTACTGGCTACTTGAACCAGACCG-3′, and 5′-TGGTTCAAGTAGCCAGTAGGACGGATGCCGAATCACTTCGTTG-3′. After ligation, DNA was transformed into competent cells, plasmid DNA was isolated from specific colonies and its sequence verified. For the construction of the mismatch bearing homologous donors, the oligonucleotides were designed to bear the specific mismatch. These were: 5′-CTGGTTCAAGTAGCCCAGGTTTTCTCAGTCACGACGGGAATACACTTCGTTG-3′ and 5′-ACCGCAACGAAGTGATTCCCGTGACTGGGAAAACCTGGGCTACTTGAACCAGACCG-3′ for the G : T mismatch; 5′-CTGGTTCAAGTAGCCCAGGTTTTCCCAGTCACGACGGGAATACACTTCGTTG-3′ and 5′-ACCGCAACGAAGTGATTCCCGTGACTGAGAAAACCTGGGCTACTTGAACCAGACCG-3′ for the A : C mismatch; 5′-CTGGTTCAAGTAGCCCAGG-TTTTCCCAGTCACGACGGGAATACACTTCGTTG-3′ and 5′-ACCGCAACGAAGTGATTCCCGTGACTGCGAAAACCTGGGCTACTTGAACCAGACCG-3′ for the C : C mismatch; and 5′-CTGGTTCAAGTAGCCCAGGTTTTCGCAGTCACGACGGGAATACACTTCGTTG-3′ and 5′-ACCGCAACGAAGTGATTCCCGTGACTGGGAAAACCTGGGCTACTTGAACCAGACCG-3′ for the G : G mismatch. After ligation to the cmR vector (150 μg), the closed circular plasmid was run on 0.8% agarose gel and the required fragment was eluted using the Elutrap electro-elution extraction protocol. This plasmid (10 μg) was then restricted with HindIII and DraI and the restriction products were once more separated on 0.8% gels and the 1072-bp segment was extracted by elutrap. The non-homologous DNA donor (nhDNA) was the 1012 SspI and HindIII restriction fragment of pCDNA3, which shares no homology with the GP.
Gapped plasmif repair assay
The in vivo assay for repair of GP by TLS and HDR is similar to the TLS assay previously described (40–42). It involves the transient transfection of the cells using the jetPEI transfection reagent (polyplus transfection, Illkirch, France) with a DNA mixture containing 50 ng of the gap-lesion plasmid (kanR), 50 ng of a normalizing GP without a lesion (cmR) and 5–10 μg of pUC18, as carrier plasmid. When assaying for HRR, 150 ng of hDNA was also introduced into the DNA mixture. Following an incubation period sufficient for gap filling (4–8 h), plasmids were extracted from the cells. Plasmid gap filling repair was assayed by introduction of the recovered DNA mixture into an E. coli recA strain, followed by plating on LB-kan and LB-cm to select for filled-in gap-lesion and normalizing plasmids, respectively. Gap-lesion repair is calculated by dividing the number of kanR colonies by the number of cmR colonies. The necessity for the use of a normalizing GP is due to the fact that the actual number of transformant colonies obtained in the different experiments varies depending on the cell line and the gap plasmid used, the gap filling efficiency, and the transformation efficiencies. Therefore, different fractions of the extracted DNA mixtures were taken to transform the indicator E. coli strain, and different volumes of transformed bacteria were plated. In order to present the data in simplified form, which allows comparison between experiment sets, the number of colonies was adjusted to a common volume of transformation mixture (100 μl). Extraction of the plasmids from the cells is conducted by alkaline conditions followed by renaturation, which allow for the recovery only of covalently closed, and thus completely filled in GPs. In this way, we assure that only the filled-in GPs recovered were indeed introduced into the bacteria cells.
To determine the DNA sequence changes that have occurred during plasmids repair, sequence analysis was carried using the TempliPhi DNA Sequencing Template Amplification Kit (GE Healthcare) and the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems). Reactions were analyzed by capillary electrophoresis on a 3130xl Genetic Analyzer (Applied Biosystems). The repair by HRR or TLS was calculated by multiplying total plasmid repair levels by the fraction of HRR or TLS events out of the total sequences analyzed.
Knocking-down the expression of RAD51
Knocking-down of RAD51 expression was conducted by transfection of SmartPool siRNA (10 nM, using Dharmafect 2, Dharmacon) to H1299 cells. Analysis of knock down was conducted by western blot with PC130 polyclonal anti-RAD51 antibody (Chalbiochem, 1 : 2500), compared to tubulin levels (mouse polyclonal antibody, Sigma, 1 : 15 000). In gap-repair experiments, the cells were transfected with siRNA, 72 h after initial transfection, cells were re-transfected and the experiment was conducted 72 h after the second siRNA boost.
RESULTS
HDR can efficiently fill in gap-lesion structures in mammalian cells
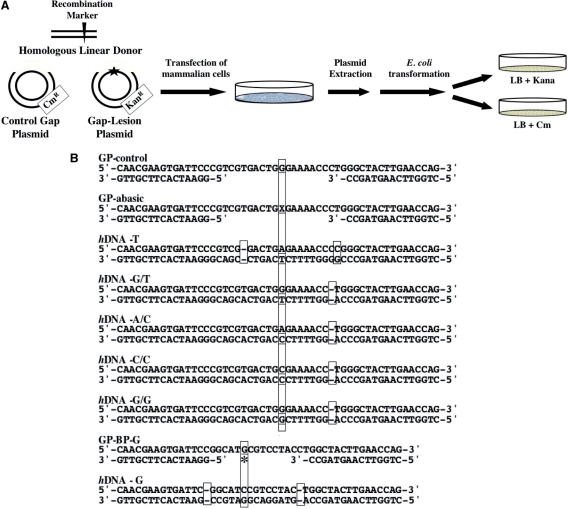
To examine whether mammalian cells can fill in DNA gaps opposite lesions, we made use of a model shuttle assay, based on a plasmid carrying a gap opposite a site-specific DNA damage (modeling a post-replication gap), and a homologous linear dsDNA (modeling a sister chromatid) (Figure 1A). The two DNAs were used to transfect cultured human cells, in which gap filling could in theory occur by TLS, or by HDR using the homologous DNA as a donor. Analysis was done by extracting the plasmid content from the mammalian cells under alkaline conditions followed by renaturation, allowing only completely repaired, covalently closed plasmids to remain intact, and using it to transform an indicator E. coli recA strain to kanamycin-resistance (the marker present on the plasmid). Plasmids were then isolated from individual colonies and subjected to DNA sequence analysis. Including in the transfection a GP without a lesion (and carrying a chloramphenicol-resistance gene) enabled determination of the efficiency of gap-lesion plasmid repair by calculating the ratio of kanR to cmR E. coli colonies. A similar assay (without the donor) has previously been useful in studying TLS in mammalian cells (39–44).
Figure 1.
A plasmid-based assay for the repair of gaps opposite lesions. (A) Outline of the experimental system. See text for details. (B) Relevant sequences of the GPs and the homologous donors used in this work. X represents the synthetic abasic site, and the star underneath the G—the benzo[a]pyrene-guanine adduct. hDNA, homologous DNA. The site of the lesion and the appropriate recombination markers are indicated by long and short vertical rectangles, respectively.
The lesion opposite the gap was a synthetic abasic site, representing one of the most common lesions in DNA. The donor homologous DNA carried a T opposite the location corresponding to the abasic site (because T is rarely inserted opposite this lesion), and two nearby markers: a −1 deletion and a single base substitution (hDNA-T; Figure 1B). This enables discrimination between gap filling by HDR (T opposite the abasic site and the nearby markers) and TLS (insertion of nucleotides or −1 deletions and no nearby markers) (Figure 1B).
Table 1 and Figure 2A describe the results of an experiment in which human H1299 cells were transfected with a mixture of a GP carrying an abasic site (kanR), the donor linear dsDNA, and the normalizing GP (cmR). Plasmid repair in the presence of the homologous donor (37%) was 3.5-fold higher than with a heterologous donor (10.7%). Consistently, 84% of the former contained the HDR markers, as opposed to none (0/54) of the latter (Table 2 and Figure 2B). Importantly, only 5% of the isolates contained large deletions, indicative of breakage of the gap-lesion plasmid, followed by NHEJ (Table 2, ‘Other events’). This indicates that when present in the mammalian cells, most of the gap-lesion substrates were not linearized by nicking at the ssDNA region. The gap filling HDR reaction was observed also with a GP carrying a site-specific bulky benzo[a]pyrene-guanine adduct (BP-G), (Tables 1 and 2 and Figure 2A and B). Interestingly, the presence of a lesion opposite the gap was required, since when the GP without a lesion was analyzed, only 2% of the isolates (1/42) carried the HRR markers (Table 2 and Figure 2B). Control experiments, in which the DNA mixture (non-denatured) was introduced directly into the E. coli indicator-strain verified that the HDR events did not occur in the indicator E. coli (Table 3). When the DNA mixture was denatured prior to E. coli transformation, extremely low transformation efficiency was observed, as expected from a recA strain (39).
Table 1.
Enhanced filling in of gaps opposite lesions in the presence of homologous DNA
| Lesion opposite the gap in the plasmid | Donor DNA |
E. coli transformantsa |
Plasmid repairb (%) | |
|---|---|---|---|---|
| KanR | CmR | |||
| Abasic site | None | 104 | 679 | 12.1 ± 3.7 |
| nhDNA | 25 | 340 | 10.7 ± 2.6 | |
| hDNA | 94 | 200 | 37 ± 7.9 | |
| BP-G | None | 47 | 648 | 5.4 ± 1.2 |
| hDNA | 89 | 258 | 15.9 ± 5.1 | |
H1299 cells were transfected with a DNA mixture containing the indicated gap-lesion plasmid (kanR) and the control gap-plasmid (cmR) in the presence or absence of a linear donor (homologous, hDNA, or non-homologous, nhDNA). Following 8 h incubation, the DNA was extracted and used to transform an E. coli indicator strain. Plasmid repair was calculated based on the ratio of kanR/cmR colonies.
aThe number of transformants obtained in a typical assay with 100 μl of transformation mixture.
bPlasmid repair values are the averages of six experiments, and therefore are not exactly equal to the ratios of KanR to CmR colonies presented in the two previous columns, which show the results of a single typical experiment.
Figure 2.
HDR repairs gaps opposite lesions in human cells. (A) Homology-dependent repair enhances repair of gaps opposite an abasic site or a BP-G adduct in the human H1299 lung cancer cell line. See Table 1 for details. (B) The percentage of repair events by HDR from the experiments depicted in (A), and presented in detail in Table 2.
Table 2.
DNA sequence analysis of gap filling opposite site-specific lesions in the presence of a homologous donor
| Gapped plasmid (GP) | GP-abasic | GP-abasic | GP-BP-G | Control GP |
|---|---|---|---|---|
| Donor DNA | nhDNA | hDNA | hDNA | hDNA |
| Nucleotide inserted opposite the lesion, or event type | ||||
| A | 10 | 1 | 1 | – |
| C | 17 | 3 | 3 | – |
| G | – | – | – | – |
| T | 1 | – | – | – |
| (−1) | 22 | 5 | – | – |
| (−2) | 1 | – | – | – |
| HDR product | 0 (<2%) | 48 (84%) | 28 (82%) | 1 (2%) |
| Control GP sequence | – | – | – | 41 |
| Other events | 3 | – | 2 | – |
| Total number of isolates | 54 | 57 | 34 | 42 |
H1299 cells were transfected with a DNA mixtures containing a plasmid with a gap opposite a synthetic abasic site (GP-abasic; kanR) or a BP-G adduct (GP-BP-G; kanR) or the control GP (cmR) in the presence of a hDNA or nhDNA, DNA partner. Following 8 h incubation, plasmids were extracted from the cells and used to transform an E. coli indicator strain. Plasmids were extracted from either kanR or cmR colonies and subjected to sequence analysis. Shown is the DNA sequence opposite the lesion obtained from individual colonies. HDR products are detected based on the nucleotide across from the lesion as well as the presence of the specific recombination markers, transferred from the hDNA sequence (Figure 1B). The single-nucleotide insertions opposite the lesions are due to TLS. For the GP-abasic and Control GP plasmids the hDNA used was hDNA-T. For the GP-BP-G it was hDNA-G. Other events are primarily large deletions.
Table 3.
DNA sequence analysis of descendants of gap-lesion plasmids that were introduced into the E. coli recA indicator strain without prior passage in mammalian cells
| Homologous donor | Number of isolates (%) |
|
|---|---|---|
| nhDNA | hDNA | |
| Nucleotide inserted opposite the abasic site | ||
| A | – | – |
| C | – | – |
| G | – | – |
| T | 1 (8) | – |
| HDR product | – | – |
| Deletions (−1) | 11 (92) | 24 (100) |
| Total number of isolates | 12 | 24 |
DNA mixtures containing the gap-lesion plasmid carrying an abasic site (kanR) together with a linear hDNA or nhDNA DNA molecule were used to directly transform the E. coli recA indicator strain. Plasmids were extracted from kanR colonies and subjected to sequence analysis. Shown is the DNA sequence opposite the lesion obtained from individual colonies. No HDR products were detected.
HDR is mediated primarily by physical transfer of the homologous DNA strand
The homologous recombination event could theoretically occur by one of two mechanisms. The complementary strand could be filled-in by direct transfer from the homologous donor, in a ‘cut and paste’ mechanism (strand transfer recombination). On the other hand, the homologous donor could serve as an alternative template for DNA synthesis, instead of the damaged template in the gap, (template switch; Figure 3A). In order to distinguish between the products of these two different mechanisms, we constructed homologous donor molecules bearing a mismatch at the position corresponding to the lesion in the GP (Figure 1B). The two donors initially used had either a G : T or an A : C mismatch (hDNA-G : T and hDNA-A : C, respectively). With the G : T mismatch, a strand transfer event, in which physical transfer occurs, would result in the occurrence of a T across from the lesion in the recovered plasmid. A template switch event, however, would involve the use of the homologue as an alternative template, insertion of a C across from the G, and a C in the product (Figure 3A). In experiments with the reciprocal A : C mismatch, a C indicates strand transfer and a T indicates a template switch event, thus ruling out a bias for either a C or a T as a product (Figure 3A).
Figure 3.
Gap-filling by HDR occurs predominantly by a strand transfer mechanism. (A) A scheme illustrating strand transfer and template switch homology-dependent repair (HDR; HRR) mechanisms for filling in gaps opposite a lesion (indicated by a star). See text for details. (B) Fraction of HDR events occurring via strand transfer (black columns) and template switch (gray columns). Shown are results in HCT116 (mismatch repair defective) and H1299 human cell lines, with a donor carrying either a G : T, A : C, C : C or G : G mismatch. Detailed data are presented in Table 4.
This assay system requires that the mismatch remain unrepaired throughout the experiment. In order to ascertain this, we conducted control experiments in which a control closed circular DNA bearing the mismatch and a cmR marker was used to transiently transfect the cells, followed by DNA extraction and introduction into a mismatch repair-deficient E. coli mutS strain. The underlying logic was that unrepaired, mismatch-bearing plasmids would yield a bacterial colony containing a mixed population of plasmids (with both a C and a T at the specific position). Sequencing of plasmids obtained from such colonies would result in both a C and a T at this position (an N in the sequence output), as was indeed found to be the case (data not shown).
When the assay was conducted with hDNA-G : T (Table 4 and Figure 3B) in H1299 cells, 24/35 sequences carried the HDR markers, of which 83% had a T, indicating a strand transfer mechanism (Table 4 and Figure 3B). Similarly, with the reciprocal hDNA-A : C, strand transfer constituted 85% of the recombination events. A similar preference for strand transfer was obtained with the human cell line HCT116, which is mismatch repair-deficient (Table 4 and Figure 3B).
Table 4.
The majority of HDR events occur through a strand transfer mechanism
| Number of isolates |
||||||
|---|---|---|---|---|---|---|
| Mismatch in donor | G : T |
A : C |
C : C |
G : G |
||
| Cell line | H1299 | HCT116 | H1299 | HCT116 | H1299 | H1299 |
| Event type | ||||||
| Base substitutions | ||||||
| A | 4 | 1 | 2 | – | 2 | 4 |
| C | 3 | – | 3 | – | – | – |
| G | – | – | 1 | – | – | – |
| T | – | – | 1 | – | – | – |
| (−1) | 3 | 5 | 9 | 1 | 1 | 2 |
| (−2) | – | – | – | – | 1 | 1 |
| T HDR | 20 | 36 | 10 | 16 | – | – |
| C HDR | 4 | 6 | 34 | 29 | 24 | 4 |
| G HDR | – | – | – | – | 7 | 37 |
| Large deletions | 1 | – | – | – | 2 | 2 |
| Total number of isolates | 35 | 48 | 60 | 46 | 37 | 50 |
DNA mixtures containing the gap plasmid carrying the abasic site (kanR) together with a homologous donor bearing either a G : T, an A : C, a C : C or a G : G mismatch at the lesion position were used to transfect the human cell lines H1299 or HCT116. Following 8 h incubation, DNA was extracted and used to transform an indicator E. coli strain. Plasmids were extracted from kanR colonies and subjected to sequence analysis. HDR products were identified based on the base at the position across from the lesion: a C or a T for the G : T and A : C mismatches, and a C or a G for the C : C and G : G mismatches, as well as the additional (−1) deletion recombination marker (Figure 1B). In the presence of a G : T mismatch, the product of a strand transfer mechanism is a T at the position across from the lesion, whereas a template switch mechanism would result in a C. In the presence of an A : C mismatch, a C indicates strand transfer whereas a T indicates template switch. With a C : C mismatch a C indicates strand transfer and a G indicates a template switch, whereas with the G : G mismatch a G indicates strand transfer whereas a C indicates template switch. The HDR strand transfer events are in bold type.
Since G:T mismatches can be repaired by a mechanism independent of the general mismatch repair system (1), and as an additional mean to rule out mismatch repair bias, we repeated similar experiments in H1299 cells with the homologous donors hDNA-C : C and hDNA-G : G, carrying a C : C or a G : G mismatch, respectively, at the position corresponding to the lesion in the GP (Figure 1B). With the C : C mismatch strand transfer is expected to yield a C opposite the location corresponding to the lesion, whereas template switch—a G. With the G : G mismatch strand transfer is expected to yield a G opposite the location corresponding to the lesion, and template switch—a C. As can be seen in Table 4 and Figure 3B, with the C : C mismatch 31/35 of the gap repair events carried the HDR markers, and 77% (24/31) of those had a C opposite the location corresponding to the lesion, indicating strand transfer HDR. With the G:G mismatch 41/48 events carried the HDR markers, and 90% (37/41) of those had a G opposite the location corresponding to the lesion, indicating strand transfer HDR. Taken together, gap repair via HDR exhibits a strong preference for a strand transfer mechanism.
RAD51 and NBS1, but not Rad18, play a role in gap filling through HRR
The major eukaryotic recombinase is the RAD51 protein, the homologue of the E. coli RecA (45). In E. coli, RecA is involved in both TLS and HDR (1). In mammalian cells, it is an essential gene and cannot be knocked out. Experiments were conducted in H1299 cells transfected with either siRNA directed against RAD51 or control siRNA treated cells. Knockdown was verified by immunobloting with an anti-RAD51 antibody (Figure 4A), and gap-lesion repair was analyzed for a gap opposite an abasic site. Treatment of the cells with siRAD51 affected viability only after the second siRNA transfection boost. This was observed in reduced plating efficiencies (about four times less cells). The experiments were conducted under the same confluence in siRAD51 and siControl treated cells. The use of a normalizing plasmid controls for all the factors but those involved in tolerating the damage. As can be seen in Table 5 and Figure 4B, HDR was reduced by 2.3-fold compared to cells treated with control siRNA. A similar effect was observed on TLS.
Figure 4.
Involvement of RAD51 and NBS1, but not Rad18, in gap-filling HDR. (A) Immunoblot analysis showing siRNA knock-down of RAD51 expression in H1299 cells either without (lanes 1 and 3) or with (lanes 2 and 4) transfection with the gapped DNAs. See methods for details. (B) Results of experiments in H1299 cells in which RAD51 was knocked-down show more then a 2-fold reduction in both HDR and TLS. (C) Experiments conducted in NBS1 cells show significant reduction in HDR without affecting TLS. (D) Experiments conducted in Rad18−/− MEFs show strong decrease in TLS in the RAD18 deficient cells with no effect on HDR. See Tables 5–7 for details.
Table 5.
RAD51 is involved in gap-lesion repair by both TLS and HDR in human cells
| Treatment | DNA substrate mix |
E. coli transformantsa |
Plasmid repair (%) | TLSb (%) | HDRb (%) | |
|---|---|---|---|---|---|---|
| KanR | CmR | |||||
| siRAD51 | GP-abasic + hDNA | 129 | 751 | 13.1 ± 5.1 | 6 ± 2.3 | 6.2 ± 2.4 |
| siControl | GP-abasic + hDNA | 187 | 460 | 29.6 ± 5.8 | 14.5 ± 2.8 | 14.5 ± 2.8 |
| Number of isolates (%) |
||
|---|---|---|
| Treatment | siRAD51 | siControl |
| Plasmid type | GP-abasic + hDNA | GP-abasic + hDNA |
| Event type | ||
| Base substitutions | ||
| A | 7 | 18 |
| C | 5 | 5 |
| G | 1 | 2 |
| T | – | – |
| (−1) deletion | 17 | 4 |
| (−2) deletion | – | – |
| Complex TLS eventsc | 4 | |
| Total TLS | 30 (46%) | 33 (49%) |
| HDR product | 31 (47%) | 33 (49%) |
| Other eventsd | 5 | 2 |
| Total number of isolates | 66 | 68 |
H1299 cells were transfected with siRNA directed against RAD51 or control siRNA. Once knock down was established, the DNA mixture containing the gap plasmid bearing an abasic site (kanR) and the control gap-plasmid (cmR) in the presence of hDNA was introduced into the cells. Following 8 h incubation, the DNA was extracted and used to transform an E. coli indicator strain. Plasmid repair was calculated based on the ratio of kanR/cmR colonies. Each result represents the average of at least four experiments. Results of single gap-filling events were obtained by sequence analysis of plasmid DNA extracted from single kanR colonies. Depicted is the sequence at the position across from the lesion.
aThe number of transformants obtained in a typical assay with 100 μl of transformation mixture.
bThe repair by HDR or TLS was calculated by multiplying total plasmid repair levels by the fraction of HDR or TLS events out of the total sequences analyzed.
cComplex TLS events involved mutation in the adjacent nucleotides. These were ACC, CAG, CCT and CCCC instead of CXC, when X represents the position across from the damage.
dOther events depict several base pair deletions or insertion of non-homologous sequence.
In order to establish whether the NBS1 gene product played a role in gap-filling recombination, we conducted similar experiments with the abasic site bearing plasmid in SV40 transformed NBS1 fibroblasts and their complemented controls. Our results show a 2.6-fold decrease in the repair of gap-lesion plasmids by HDR in NBS1 deficient cells, while TLS was unaffected (Table 6 and Figure 4C). In contrast, experiments performed in Rad18 knockout MEFs compared to wild type controls showed that the Rad18 gene did not affect gap-filling HDR, while it was required for TLS (Table 7 and Figure 4D).
Table 6.
NBS1 is involved in gap-lesion repair by HDR but not TLS in human cells
| Cell line |
E. coli transformantsa |
Plasmid repair (%) | TLSb (%) | HDRb (%) | |
|---|---|---|---|---|---|
| KanR | CmR | ||||
| Transformed NBS1−/− | 182 | 635 | 34.6 ± 10.1 | 12.5 ± 3.6 | 13.1 ± 3.6 |
| Transformed Comp. | 109 | 185 | 49.7 ± 7.5 | 11.9 ± 3.8 | 34.2 ± 10.9 |
| Number of isolates (%) |
||
|---|---|---|
| Cell line | NBS1−/− | NBS1-complemented |
| Plasmid type | GP-abasic + hDNA | GP-abasic + hDNA |
| Event type | ||
| Base substitutions | ||
| A | 10 | 7 |
| C | 2 | 1 |
| G | – | – |
| T | 3 | 2 |
| HDR product | 18 (38%) | 37 (69%) |
| Deletions | – | – |
| (−1) | 2 | 3 |
| Other eventsc | 12 | 4 |
| Total number of isolates | 47 | 54 |
Transformed GM07166 NBS1 cells and their complemented controls were transfected with DNA mixture containing the gap plasmid bearing an abasic site (kanR) and the control gap-plasmid (cmR) in the presence of homologous DNA (hDNA). Following 4 h incubation, the DNA was extracted and used to transform an E. coli indicator strain. Plasmid repair was calculated based on the ratio of kanR/cmR colonies. Each result represents the average of four experiments. Results of single gap-filling events were obtained by sequence analysis of plasmid DNA extracted from single kanR colonies. Depicted is the sequence at the position across from the lesion.
aThe number of transformants obtained in a typical assay with 100 μl of transformation mixture.
bThe repair by HDR or TLS was calculated by multiplying total plasmid repair levels by the fraction of HDR or TLS events out of the total sequences analyzed.
cOther events depict several base pair deletions or insertion of non-homologous sequences.
Table 7.
Rad18 is involved in gap-lesion repair by TLS but not in HDR
| Cell line |
E. coli transformantsa |
Plasmid repair (%) | TLSb (%) | HDRb (%) | |
| KanR | CmR | ||||
| Rad18−/− | 75 | 320 | 23.8 ± 10.6 | <1.2 ± 0.5 | 22.6 ± 10 |
| Rad18+/+ | 78 | 140 | 58.4 ± 9 | 21.6 ± 3.3 | 27.4 ± 4.3 |
| Number of isolates (%) |
||
| Cell line | Rad18−/− | Rad18+/+ |
| Plasmid type | GP-BPG + hDNA | GP-BPG + hDNA |
| Event type | ||
| Base substitutions | ||
| A | – | 2 |
| C | – | 4 |
| G | – | 1 |
| T | – | – |
| (−1) | – | – |
| Total TLS | 0 (<5%) | 7 (37%) |
| Control GP sequence | 1 | 1 |
| HDR product | 18 (95%) | 9 (47%) |
| Other eventsc | – | 2 |
| Total number of isolates | 19 | 19 |
Rad8−/− and Rad18+/+ MEF cells were transfected with DNA mixture containing the gap plasmid bearing a BP-G adduct (kanR) and the control gap-plasmid (cmR) in the presence of hDNA. Following 10 h incubation, the DNA was extracted and used to transform an E. coli indicator strain. Plasmid repair was calculated based on the ratio of kanR/cmR colonies. Each result represents the average of at least four experiments. Results of single gap-filling events were obtained by sequence analysis of plasmid DNA extracted from single kanR colonies. Depicted is the sequence at the position across from the lesion.
aThe number of transformants obtained in a typical assay with 100 μl of transformation mixture.
bThe repair by HDR or TLS was calculated by multiplying total plasmid repair levels by the fraction of HDR or TLS events out of the total sequences analyzed.
cOther events depict several base pair deletions or insertion of non homologous sequences.
DISCUSSION
There is accumulating evidence that ssDNA gaps are formed in a variety of cells following treatment with DNA-damaging agents, including E. coli, S. cerevisiae and human cells (2–4,46). These gaps can be filled by either TLS or HDR. Based on the S. cerevisiae paradigm (47), two HDR pathways have been proposed, which differ by the mode by which the homologous donor (usually a sister chromatid) is used to fill in the gap opposite the lesion. In strand transfer HDR the gap is filled by physical transfer of the complementary strand from the donor, whereas in template-switch HDR the missing segment is copied from the donor. Gap filling tolerance by HDR—be it via strand transfer or template switch mechanisms—offers a potential advantage over TLS in being inherently accurate, in contrast to the inherently error-prone nature of TLS. Yet, while the operation of TLS is well established in mammalian cells, the operation of the two HDR mechanisms in filling in gaps opposite lesions has been more difficult to pin down.
The use of the plasmid-based shuttle assay system, which contains a defined model HDR substrate, enabled us to establish, for the first time, that gaps opposite lesions can be accurately filled in by HDR in mammalian cells in general, and human cells in particular. Our data indicate that in this system the events detected in the E. coli indicator strain have occurred in the mammalian cells, and not in the indicator bacterial cells. This conclusion is supported by the following arguments: (i) the E. coli strain used was RecA-deficient and therefore defective in both HRR and TLS across abasic sites and BP-G adducts (1). (ii) The extraction of the plasmid from the mammalian cells was done using a protocol of alkaline denaturation followed by renaturation. Under such conditions all gapped or nicked plasmids remain denatured, and poorly transform the indicator E. coli strain. Only plasmids that have been fully filled in and ligated in the mammalian cells remain covalently closed and were able to transform the indicator E. coli cells. (iii) Transformation of the E. coli cells without prior passage in the mammalian cells yielded a background and extremely low number of colonies. (iv) DNA sequence of plasmids from transformants that were obtained from E. coli cells without prior passage through mammalian cells did not show any of the markers of HDR.
Our conclusion that the gap filling events that we have observed in the mammalian cells represent HDR are supported by their dependence on a homologous donor, on the dependence on a DNA lesion opposite the gap in the acceptor, on the presence of the donor markers in the descendents of the filled in gap-lesion plasmids, and by the effects on gap-filling in cells deficient in genes related to TLS and HDR.
Based on the S. cerevisiae paradigm the Rad18 ubiquitin ligase controls both TLS and template-switch HDR, but not strand transfer HDR, whereas the Rad51 recombination protein is involved both modes of HDR (1,47). Consistently, gap filling by HDR in our assay was decreased in a human cell line in which the expression of RAD51 was knocked-down, but not in a Rad18−/− MEF, compared to their ‘wild-type’ counterparts. Of note are the contrasting effects observed in NBS1−/− and Rad18−/− mutant cells: whereas the former affected HDR but not TLS, the latter affected TLS but not HDR. The function of the NBS1 protein is not fully understood, but its involvement in gap filling HDR is consistent with the finding that XRS2, the yeast homologue of NBS1, is involved in the two HDR pathways (47).
RAD51 is the eukaryotic homologue of the E. coli recA, which is known to be involved in bacterial DNA damage tolerance through both TLS and HRR (1). In mammalian cell, however, RAD51 is an essential gene and cannot be knocked out (48,49). We therefore resorted to knocking down the expression of RAD51 using siRNA. Under conditions where RAD51 expression was effectively knocked-down (∼95%), gap filling was reduced by 2.3-fold compared to cells treated with a control siRNA. This rather moderate effect might be an underestimation of the RAD51 dependence in chromosomal gap filling by HDR, and may stem from the use of the plasmid model assay system, and gaps much shorter than chromosomal gaps. Under these conditions, either residual RAD51 activity, or the activity of homologous proteins with redundant function, may be responsible for partially promoting HDR. Nevertheless, these results suggest that RAD51 is involved in the gap-filling reaction through both HDR and TLS. A role of RAD51 in mammalian TLS was not previously reported, and warrants further investigation, however, it is noteworthy that human RAD51 was reported to interact with DNA polymerase η, one of the major TLS polymerases (50). This resembles the situation in E. coli, where RecA is required for both TLS and HDR, suggesting functional evolutionary conservation.
The design of a donor DNA with a mismatch at the location corresponding to the damaged site in the acceptor plasmid enabled us to assay simultaneously strand transfer and template switch mechanisms of HDR. Our results indicate that the majority of gap filling events had occurred via a strand transfer mechanism. We take these results to indicate that there are situations in the cell under which strand transfer predominates over template switch as the HDR gap repair mechanism, e.g. the filling in of post-replication gaps. This is similar to the results obtained with a plasmid system in E. coli (29,30) and with the Howard–Flanders model for HRR in UV irradiated E. coli cells (46,51). However, there might exist situations in which template switch will be preferred. For example, when replication forks encounter DNA lesions template switch may be favored over strand transfer as a damage tolerance mechanism (52). In addition, there might be a discrimination between the leading and lagging strands, with template switch favored on the former, and strand transfer on the latter, as suggested for S. cerevisiae (47).
In summary, our results indicate, for the first time, that mammalian cells can repair gaps opposite lesions by HDR, acting primarily by a strand transfer mechanism. This mechanism involves RAD51 and NBS1, but not Rad18. Additional studies assaying directly chromosomal HDR are needed to examine whether such mechanisms operate in the context of mammalian chromosomes.
FUNDING
Flight Attendant Medical Research Institute, Florida, USA; the Israel Science Foundation (no. 564/04 and 1136/08 to Z.L.); the National Institutes of Health, USA (no. CA099194 to N.G.); Z.L. is the incumbent of the Maxwell Ellis Professorial Chair in Biomedical Research. Funding for open access charge: Flight Attendant Medical Research Institute, Florida, USA.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Yosef Shiloh (Tel Aviv University, Israel) for NBS1 and complemented cells, and Satoshi Tateishi (Kumamoto University, Japan) for Rad18−/− and Rad18+/+ MEF.
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. [Google Scholar]
- 2.Lehmann AR, Fuchs RP. Gaps and forks in DNA replication: rediscovering old models. DNA Repair. 2006;5:1495–1498. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Mojas N, Lopes M, Jiricny J. Mismatch repair-dependent processing of methylation damage gives rise to persistent single-stranded gaps in newly replicated DNA. Genes Dev. 2007;21:3342–3355. doi: 10.1101/gad.455407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl Acad. Sci. USA. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Livneh Z. Keeping mammalian mutation load in check: regulation of the activity of error-prone DNA polymerases by p53 and p21. Cell Cycle. 2006;5:1918–1922. doi: 10.4161/cc.5.17.3193. [DOI] [PubMed] [Google Scholar]
- 7.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 8.Zheng H, Wilson JH. Gene targeting in normal and amplified cell lines. Nature. 1990;344:170–173. doi: 10.1038/344170a0. [DOI] [PubMed] [Google Scholar]
- 9.Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc. Natl Acad. Sci. USA. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorrell DA, Kolb AF. Targeted modification of mammalian genomes. Biotechnol. Adv. 2005;23:431–469. doi: 10.1016/j.biotechadv.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Godwin AR, Bollag RJ, Christie DM, Liskay RM. Spontaneous and restriction enzyme-induced chromosomal recombination in mammalian cells. Proc. Natl Acad. Sci. USA. 1994;91:12554–12558. doi: 10.1073/pnas.91.26.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shulman MJ, Collins C, Connor A, Read LR, Baker MD. Interchromosomal recombination is suppressed in mammalian somatic cells. EMBO J. 1995;14:4102–4107. doi: 10.1002/j.1460-2075.1995.tb00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benjamin MB, Potter H, Yandell DW, Little JB. A system for assaying homologous recombination at the endogenous human thymidine kinase gene. Proc. Natl Acad. Sci. USA. 1991;88:6652–6656. doi: 10.1073/pnas.88.15.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramani S, Rubnitz J. Recombination events after transient infection and stable integration of DNA into mouse cells. Mol. Cell Biol. 1985;5:659–666. doi: 10.1128/mcb.5.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cahill D, Connor B, Carney JP. Mechanisms of eukaryotic DNA double strand break repair. Front Biosci. 2006;11:1958–1976. doi: 10.2741/1938. [DOI] [PubMed] [Google Scholar]
- 16.Pastink A, Eeken JC, Lohman PH. Genomic integrity and the repair of double-strand DNA breaks. Mutat. Res. 2001;480–481:37–50. doi: 10.1016/s0027-5107(01)00167-1. [DOI] [PubMed] [Google Scholar]
- 17.Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutat Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 19.Tashiro S, Kotomura N, Shinohara A, Tanaka K, Ueda K, Kamada N. S phase specific formation of the human Rad51 protein nuclear foci in lymphocytes. Oncogene. 1996;12:2165–2170. [PubMed] [Google Scholar]
- 20.Hendrickson EA. Cell-cycle regulation of mammalian DNA double-strand-break repair. Am. J. Hum. Genet. 1997;61:795–800. doi: 10.1086/514895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Essers J, van Steeg H, de Wit J, Swagemakers SM, Vermeij M, Hoeijmakers JH, Kanaar R. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 2000;19:1703–1710. doi: 10.1093/emboj/19.7.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair. 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Helleday T, Arnaudeau C, Jenssen D. Effects of carcinogenic agents upon different mechanisms for intragenic recombination in mammalian cells. Carcinogenesis. 1998;19:973–978. doi: 10.1093/carcin/19.6.973. [DOI] [PubMed] [Google Scholar]
- 25.Helleday T, Nilsson R, Jenssen D. Arsenic[III] and heavy metal ions induce intrachromosomal homologous recombination in the hprt gene of V79 Chinese hamster cells. Environ. Mol. Mutagen. 2000;35:114–122. doi: 10.1002/(sici)1098-2280(2000)35:2<114::aid-em6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 26.Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward JD, Barber LJ, Petalcorin MI, Yanowitz J, Boulton SJ. Replication blocking lesions present a unique substrate for homologous recombination. EMBO J. 2007;26:3384–3396. doi: 10.1038/sj.emboj.7601766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tourriere H, Pasero P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair. 2007;6:900–913. doi: 10.1016/j.dnarep.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Berdichevsky A, Izhar L, Livneh Z. Error-free recombinational repair predominates over mutagenic translesion replication in E. coli. Mol. Cell. 2002;10:917–924. doi: 10.1016/s1097-2765(02)00679-2. [DOI] [PubMed] [Google Scholar]
- 30.Izhar L, Goldsmith M, Dahan R, Geacintov N, Lloyd RG, Livneh Z. Analysis of strand transfer and template switching mechanisms of DNA gap repair by homologous recombination in Escherichia coli: predominance of strand transfer. J. Mol. Biol. 2008;381:803–809. doi: 10.1016/j.jmb.2008.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozgenc AI, Szekeres ES, Lawrence CW. In vivo evidence for a recA-independent recombination process in Escherichia coli that permits completion of replication of DNA containing UV damage in both strands. J. Bacteriol. 2005;187:1974–1984. doi: 10.1128/JB.187.6.1974-1984.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl Acad. Sci. USA. 2005;102:15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res. 2001;477:131–153. doi: 10.1016/s0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 34.Kawabata M, Kawabata T, Nishibori M. Role of recA/RAD51 family proteins in mammals. Acta Med. Okayama. 2005;59:1–9. doi: 10.18926/AMO/31987. [DOI] [PubMed] [Google Scholar]
- 35.Brower M, Carney DN, Oie HK, Gazdar AF, Minna JD. Growth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res. 1986;46:798–806. [PubMed] [Google Scholar]
- 36.Brattain MG, Levine AE, Chakrabarty S, Yeoman LC, Willson JK, Long B. Heterogeneity of human colon carcinoma. Cancer Metastasis Rev. 1984;3:177–191. doi: 10.1007/BF00048384. [DOI] [PubMed] [Google Scholar]
- 37.Tauchi H, Kobayashi J, Morishima K, Matsuura S, Nakamura A, Shiraishi T, Ito E, Masnada D, Delia D, Komatsu K. The forkhead-associated domain of NBS1 is essential for nuclear foci formation after irradiation but not essential for hRAD50[middle dot]hMRE11[middle dot]NBS1 complex DNA repair activity. J. Biol. Chem. 2001;276:12–15. doi: 10.1074/jbc.C000578200. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avkin S, Adar S, Blander G, Livneh Z. Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc. Natl Acad. Sci. USA. 2002;99:3764–3769. doi: 10.1073/pnas.062038699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase kappa. J. Biol. Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 41.Avkin S, Sevilya Z, Toube L, Geacintov N, Chaney SG, Oren M, Livneh Z. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol. Cell. 2006;22:407–413. doi: 10.1016/j.molcel.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 42.Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adar S, Livneh Z. Translesion DNA synthesis across non-DNA segments in cultured human cells. DNA Repair. 2006;5:479–490. doi: 10.1016/j.dnarep.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Hendel A, Ziv O, Gueranger Q, Geacintov N, Livneh Z. Reduced efficiency and increased mutagenicity of translesion DNA synthesis across a TT cyclobutane pyrimidine dimer, but not a TT 6-4 photoproduct, in human cells lacking DNA polymerase eta. DNA Repair. 2008;7:1636–1646. doi: 10.1016/j.dnarep.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 46.Rupp WD, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 47.Gangavarapu V, Prakash S, Prakash L. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell Biol. 2007;27:7758–7764. doi: 10.1128/MCB.01331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 50.McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 51.Rupp WD, Wilde III CE, Reno DL, Howard-Flanders P. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J. Mol. Biol. 1971;61:25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- 52.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]