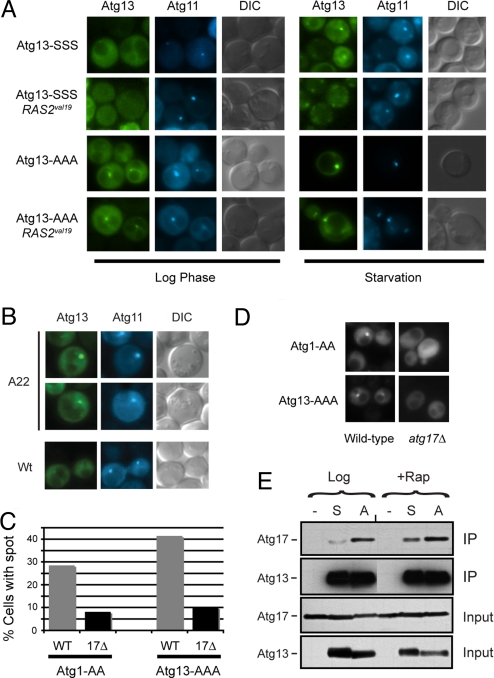
Fig. 4.
The PAS localization of Atg13 was regulated by PKA phosphorylation. (A) Fluorescence microscopy was performed with cells that contained the indicated YFP-Atg13 fusion proteins. The Atg13 proteins had either wild-type (Atg13-SSS) or nonphosphorylatable versions (Atg13-AAA) of the three PKA sites. The CFP-Atg11 fusion protein was present in all cells and served as a marker for the PAS. The RAS2val19 allele was present in the indicated strains. Nitrogen starvation was achieved by transferring the cells from SC glucose minimal medium to the SD-N medium for 1 h at 30 °C. (B) The localization of a wild-type Atg13-YFP protein in the indicated cells was assessed by fluorescence microscopy. A22, RAS2ala22. (C) Atg17 was required for the efficient localization of Atg13-AAA to the PAS in log phase cells. The fraction of cells with Atg1-AA or Atg13-AAA present in a perivacuolar punctate spot in the indicated strains is shown (20). At least 50 cell images were examined for each strain. (D) Representative images for the indicated strains in C. (E) Alteration of the PKA phosphorylation sites in Atg13 influenced the interaction with Atg17. The indicated Atg13 proteins were immunoprecipitated from yeast cell extracts and the relative levels of the associated Atg17 were assessed by Western blotting. The cell extracts were prepared from either mid-log phase cultures (Log) or from cells that were treated with rapamycin (Rap). S, Atg13-SSS; A, Atg13-AAA.