Abstract
“Chromosome conformation capture” (3C) is a powerful method to detect physical interaction between any two genomic loci. 3C involves formaldehyde crosslinking to stabilize transient interactions, followed by restriction digestion, ligation and locus-specific PCR. Accordingly, 3C reveals complex three-dimensional interactions between distal genetic elements within intact cells at high resolution. Here, we describe a modified 3C protocol designed for detection of transient chromatin interactions in the yeast Saccharomyces cerevisiae. Using this protocol, we are able to detect juxtaposition of promoter and terminator regions of genes with ORFs as short as 1 kb in length. We anticipate that this method will be generally applicable to detect dynamic, short-range chromatin interactions and will facilitate the characterization of gene loops and their functional consequences.
Keywords: chromatin, transcription, gene loop, promoter, terminator
1. Introduction
Extraordinary progress has been made in the past 25 years toward understanding the structure of the eukaryotic genome and the mechanisms that underlie gene expression. Many of the proteins required for accurate and regulated transcription have been identified and structures for many of these proteins, including RNA polymerase II, have been solved at atomic resolution (1). The structure of the nucleosome has also been solved at high resolution and the protein complexes that modify and remodel chromatin during the transcription cycle have been defined.
Missing from the picture, though, is a fundamental understanding of the three-dimensional organization of the genome and how higher order chromatin structure affects gene expression. Until recently, chromosome organization was defined primarily by microscopic techniques at low resolution (2). With the advent of new technologies, however, chromatin loops that juxtapose distal regions of the genome have been identified as important regulators of eukaryotic gene expression (3–7). Loops can serve to insulate chromatin from neighboring domains or to cluster genes with similar patterns of expression at discrete transcriptional “hubs.” Thus, chromatin architecture that juxtaposes distal regulatory elements with their cognate genes, often located tens of kilobase pairs away, is emerging as a fundamental mechanism controlling gene expression.
A key technological innovation that has allowed for detection and characterization of long range chromatin interaction is called “chromosome conformation capture” (3C) (8). According to 3C, transient chromatin interactions are stabilized by formaldehyde crosslinking, followed by extraction and digestion with a specific restriction enzyme. DNA fragments are then ligated in dilute solution under conditions that favor intramolecular ligation. Following reversal of crosslinks, ligation products are detected by PCR (Fig. 1). Thus, 3C converts physical chromatin interactions into specific ligation products with the abundance of PCR products representing the frequency of interaction. 4C, 5C and 6C derivatives of 3C were subsequently developed for high-throughput, genome-wide analyses of chromatin architecture and for identification proteins that might facilitate these higher order chromatin conformations (9–15).
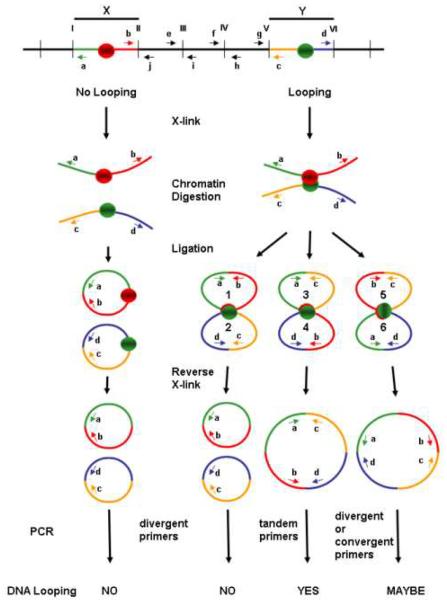
Fig. 1. Schematic depiction of the outcome of 3C analysis using divergent, convergent and tandem primer pairs.
Primers are denoted by lower case letters a – j. Restrictions sites are denoted by roman numerals I – VI. DNA fragments X and Y are either juxtaposed such that crosslinking occurs between the regions depicted as red and green circles (right side) or are not juxtaposed and do not become crosslinked (left side). PCR products from tandem primer pairs a &c or b & d are definitive for juxtaposition of fragments X and Y, whereas PCR products from divergent primer pairs a & d are definitive for X–Y juxtaposition only if PCR controls, e.g., using convergent primer pairs b & j, e & i, f & h or g &c, are included to established cutting of DNA between fragments X and Y. PCR products from convergent primer pairs b & c are definitive for looping, but should be avoided because of potential to yield multiple PCR products and to deplete PCR reactions of primers and dNTP substrates.
Protocols have been published for detection of chromatin loops by 3C technology in metazoan organisms (16–18) and for detection of long-range loops in yeast (8, 19, 20). In the yeast S. cerevisiae, enhancer-like upstream activation sequences (UAS) are generally very close (<500 bp) to their target promoters and most genes lack introns. Accordingly, yeast genes and their regulatory sequences span relatively short distances, typically not more than a few thousand base pairs. Therefore, 3C protocols that were designed to detect long range chromosomal interactions are not necessarily adequate to detect gene loops in yeast. Here we describe a highly efficient adaptation of 3C that detects dynamic, short-range chromatin interactions, including promoter-terminator juxtaposition.
2. Experimental design
There are two key features of our 3C method for detection of short-range chromatin interactions. One is formaldehyde crosslinking in whole cells, rather than isolated nuclei, a condition that we find to be essential for stabilization of transcription-dependent chromatin interactions. The second is extensive restriction digestion of chromatin samples, in effect assuring that promoter and terminator regions are untethered in the absence of crosslinking. Using our method, we have detected physical interactions between promoter and terminator regions for genes with ORFs as short as 1 kb in length (21, 22). We present our 3C protocol, including critical controls, and discuss considerations for optimizing the assay. We anticipate that this protocol will serve as a convenient method to define the structural requirements for the formation and persistence of gene loops; to assess the physiological importance of these structures in controlling gene expression; and to assess whether co-regulated genes might cluster into regulatory hubs, perhaps akin to the transcriptional hubs in mammalian cells.
2.1. Yeast strains
We have successfully used 3C to detect gene loops using derivatives of commonly used laboratory strains, including the BY4741 deletion set (Invitrogen), S288c (23), and W303 (24). Detection of gene loops by 3C in wild type strains is unaffected by elevated growth temperature; thus, loops can be detected with equal efficiency in cells incubated at 30°C or 37°C. This observation is important because it allows for the use of temperature-sensitive mutants to identify looping requirements. For example, we were able to demonstrate that looping is transcription-dependent by observing diminished looping in a temperature-sensitive rpb1-1 mutant at 37°C; the critical control showed that looping was unaffected at 37°C in the wild type strain (21, 22).
2.2. Choice of genes and restriction enzymes
The foremost consideration in choosing genes for analysis by 3C is the distribution of appropriate restriction sites. Restriction sites must flank the putative interacting regions, designated fragments X and Y in Fig. 1. Keep in mind that the closer the restriction sites are to the region of interest, the greater the degree of resolution by 3C. If working with divergent primers (see “PCR primer design” below), it is essential that at least one restriction site, and preferably multiple sites, occur between the fragments of interest in order to demonstrate that ligation products are crosslink-dependent.
More than a single restriction enzyme can also be used if these enzymes generate compatible cohesive ends for subsequent ligation. For example, BamHI, BclI and BglII can be used simultaneously as all three enzymes generate identical GATC cohesive ends. Restriction enzymes with four-base pair recognition sites are likely to yield higher 3C resolution as these enzymes cut the genome on average every 256 base pairs, although enzymes that generate four-base cohesive ends should be used to facilitate the subsequent ligation step. Restriction enzymes that cut at 65°C cannot be used in 3C analysis of yeast because prolonged incubation at 65°C disrupts DNA-protein crosslinking (25).
2.3. PCR primer design
There are several important issues to consider when designing 3C primer pairs. Primer length should be typically 20–30 bp with 40–50% GC content and not more than 5°C difference in melting temperature among the primers. Each primer should be analyzed to confirm that its sequence is unique in the genome. New primers should be designed if multiple PCR products are observed. The size of PCR products should be relatively small (150–350 bp), allowing for detection of quantitative differences between PCR products. Primers should be positioned 50–200 bp from the adjacent restriction sites (Fig. 1).
The orientation of PCR primers is an important consideration. Primers pairs can be either tandem or divergent, for example, divergent primer pairs a & d, or tandem primer pairs a & c or b & d (Fig. 1). Generation of PCR products from divergent primer pairs, however, can occur without the DNA template necessarily having been cut between the primer pairs. For example, DNA digested at sites I and VI, but not at sites II through V, could be ligated to yield PCR products using primers a & d (Fig. 1). Accordingly, PCR products from primers a & d do not conclusively reflect crosslink-dependent ligation reactions, but could represent “intramolecular” ligation between distal ends I and VI. Although the probability of DNA being cut at sites I and VI and not at intervening sites decreases as the number of internal cut sites increases, the possibility that the DNA has not been cut between divergent primer pairs must be considered, given the sensitivity of the 3C PCR assay. Accordingly, meaningful 3C data using divergent primer pairs requires that control PCR reactions be performed using convergent primer pairs to monitor digestion at internal cut sites (Fig. 2) (see also Ref.(22)).
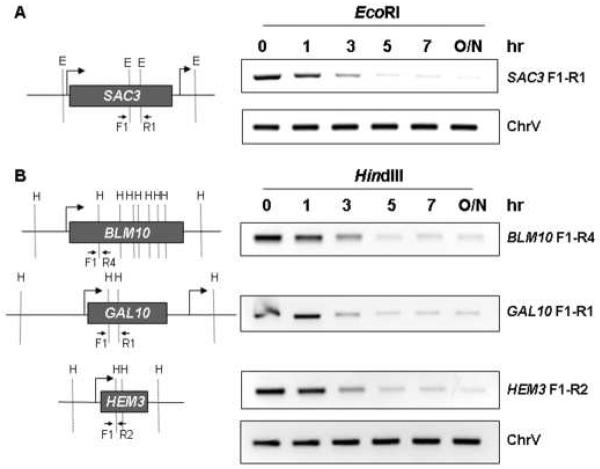
Fig. 2.
Kinetics of restriction digestion of crosslinked chromatin using either EcoRI (panel A) or HindIII (panel B). The SAC3, BLM10, GAL10 and HEM3 genes are depicted on the left side, along with the positions of the EcoRI (E) or HindIII (H) restriction sites, along with primer pairs used for convergent PCR, drawn approximately to scale. The panels on the right show the kinetics of restriction digestion of chromatin DNA, processed according to our 3C protocol (without the ligation step). Following incubation with restriction enzyme for the indicated number of hours (hr) or overnight (O/N), digestion was monitored by PCR using the convergent primer pairs depicted in the panels on the left. Chr V is a PCR input control corresponding to an intergenic region of chromosome V. All primer pairs are defined in Supplemental Data, Table S1.
Tandem primer pairs circumvent this issue and are therefore preferable to divergent primer pairs. For example, primer pairs b & d, or a & c, are preferable to primer pairs a & d because PCR products of the correct size can be generated only if the DNA has been cut between fragments X and Y (Fig. 1). In other words, tandem primer pairs eliminate concern over PCR products resulting from crosslink-independent ligation. Convergent primer pairs, e.g., b & c, would also eliminate background from intramolecular ligation, but should be avoided because annealing of these primers with template DNA that is uncut between fragments X and Y have the potential to yield multiple PCR products and to deplete PCR reactions of primers and dNTP substrates.
2.4. Controls
Generation of meaningful 3C data is critically dependent upon proper controls. These were summarized previously (19) and are discussed below in the context of our 3C protocol.
2.4.1. Primer pair efficiency
When comparing 3C data using multiple PCR primers, it is critical to control for primer pair efficiency. Template DNA that serves as a control for primer pair efficiency can readily be generated by digesting and ligating PCR-amplified DNA encompassing the region of interest (Fig. 3C, right panel). Quantitative data can then be obtained by normalizing 3C PCR products with PCR products from the randomly ligated DNA. Control template DNA can also be generated by digestion and random ligation of purified genomic DNA, although template DNA generated in this manner is more likely to include random insertions of genomic DNA between the restriction fragments to which the primers anneal. In our hands, control PCR products generated from template DNA derived from total genomic DNA often includes the anticipated product as the smallest PCR products, as well as multiple, larger products.
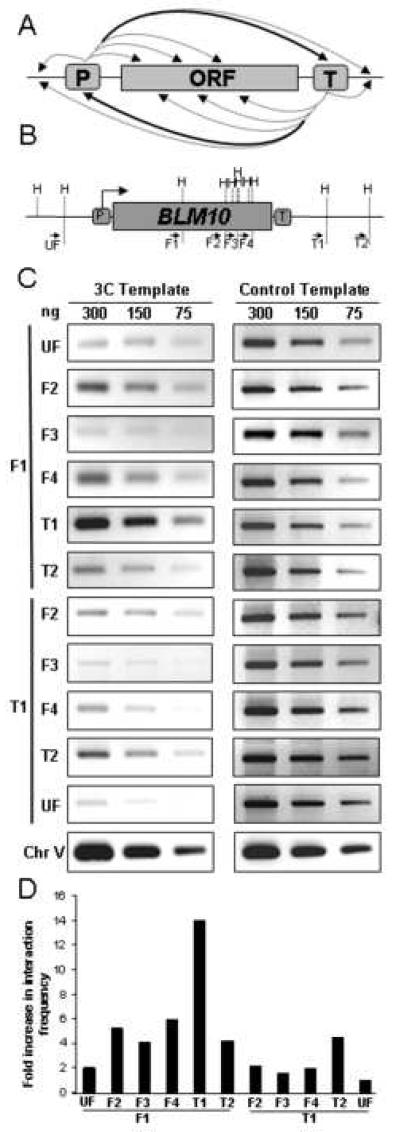
Fig. 3. Mapping short range interactions.
(A) General strategy for mapping juxtaposed regions in gene loops. As depicted here, 3C primer pairs are designed to detect juxtaposition of promoter (P) or terminator (T) regions with regions of the open reading frame (ORF), as well as upstream and downstream regions. The heavy arrows denote interactions detected by 3C for the BLM10 gene (panel C) and other genes (21, 22), whereas the dotted arrows denote 3C interactions that are not above background. (B) Schematic depiction of the BLM10 gene, depicting the positions of the HindIII sites (H) and 3C primer pairs (solid arrows). Spacing is drawn approximately to scale. (C) PCR products using the indicated primer pairs were fractionated in a 1.5% agarose gel and visualized by ethidium bromide staining using an AlphaImager 2000. Panels on the left represent 3C output analysis using the indicated concentrations of DNA template. Panels on the right correspond to PCR output from 3C control template DNA. Chr V denotes a non-transcribed region of chromosome V, generated using convergent primer pairs that are not separated by a HindIII restriction site. (D) Quantification of 3C PCR data from panel C, normalized to the 3C T1-UF PCR signal.
2.4.2. Random interactions
Foremost, it is essential to show that 3C PCR products are dependent upon formaldehyde crosslinking: PCR products reflective of looping should be significantly diminished in control samples not treated with formaldehyde (21). Second, control samples should also establish that no PCR products are detected in the absence of ligation. Third, to conclude from 3C data that DNA loops juxtapose distal regions of the gene, it is essential to demonstrate that these two regions do not randomly associate with DNA along the entire length of the gene. This issue can be addressed by “walking” along the gene using one common primer paired with primers adjacent to internal restriction sites. For example, if fragments X and Y are juxtaposed as a consequence of looping, then primer pairs b & f, and b & g should yield significantly less PCR product than primer pairs b & d (Fig. 1). Similarly, primer pairs c & i, and c & j should yield less PCR product than primer pairs c & a. We consistently observe that promoter and terminator primers paired with internal primers yield consistently less PCR product than promoter-terminator primer pairs (Fig. 3) (21, 22). Primer pairs from adjacent DNA fragments (e.g., primers b & e or primers c & h) can yield relatively high levels of PCR products, presumably due to crosslinking of proximal DNA (e.g., proximal nucleosomes) that is independent of looping.
2.4.3. Strain comparisons
Comparison of 3C data between two different strains requires data normalization. This can be done by PCR amplification of input control template DNA. In our experiments, we usually amplify a non-transcribed region of chromosome V employing the same primer pair used in our chromatin immunoprecipitation experiments (21, 22). 3C PCR data is then normalized to this input control PCR product. Accordingly, 3C data can be directly compared among different strains or different growth conditions.
2.4.4. Linearity
3C is designed such that PCR levels reflect the amount of ligated template DNA as a measure of the frequency of physical interaction between specific regions of the genome. This requires that template DNA be titrated to determine the linear range of PCR amplification. The most reliable method to quantify PCR data is to use real-time PCR, which assures that data is obtained in the linear range where PCR products reflect DNA template concentrations (19). Reliable quantitative data can also be generated by standard PCR where products are visualized in an agarose gel and quantified using a PhotoImager (Fig. 3). Alternatively, PCR amplification can be performed in the presence of a radioactive substrate (e.g., α-[32P]dATP); radioactive PCR products are then resolved in 8% polyacrylamide-TBE gels and quantified using a PhosphorImager.
3. Step-by-step protocol
3.1. Cell culture
Inoculate a 5 ml seed culture in appropriate growth medium from a freshly growing colony on a Petri dish. Recipes for commonly used yeast media are described elsewhere (26). Incubate overnight, with shaking, at 30°C. From the overnight culture, inoculate 50 ml of growth medium in a 250 ml flask to OD600~0.15. Incubate cells with shaking at 30°C until they reach an OD600 of 0.65 to 0.8. For a wild type strain growing in rich medium at 30°C this will take approximately 5 hr. When using temperature-sensitive mutants, harvest cells at mid-log phase and resuspend in pre-warmed (37°C) medium, followed by incubation at 37°C for 1 hr. If the cell density is higher than 0.8 (OD600~0.9–1.0), use less (e.g. 20 μl) chromatin for restriction digestion. Be careful to harvest different strains, or different cultures of the same strain, at the same OD600.
3.2. Chromatin crosslinking
Add 1.4 ml of 37% formaldehyde (final concentration = 1%) to 50 ml culture. Mix thoroughly and incubate 15 – 25 min at room temperature, either on a shaker or by swirling briefly every 5 min. For detection of short range gene loops it is important that the time of crosslinking not exceed 20 – 25 min. We are able to detect gene loops that specifically juxtapose 5' and 3' ends at HEM3 (984 bp ORF) following 20 min of crosslinking (22). Longer periods of exposure to formaldehyde result in enhanced crosslinking along the length of the gene.
Add 5.4 ml of 1.25 M glycine to quench excess formaldehyde. Mix and incubate for 5 min at room temperature either on shaker or by swirling gently every 1 min. Transfer cells to a 50 ml centrifuge tube. Pellet cells by centrifugation at 3,000 × g for 10 min at 4°C using a Sorvall SH3000 centrifuge or equivalent. Wash cells once with 10 ml of ice cold TBS containing 1% Triton × 100. Pellet cells and resuspend in 1 ml of ice cold TBS containing 1% Triton × 100 and transfer to a 1.5 ml Eppendorf tube. Pellet cells by centrifugation at 10,000 g for 10 min at 4°C in a Microcentrifuge, then aspirate buffer. Resuspend the pellet thoroughly in 500 μl of FA lysis buffer by pipeting up and down. Freeze cells in liquid nitrogen and store at −80°C. Cells can be stored at this point at −80°C for up to one year.
3.3. Isolation of crosslinked chromatin
Thaw cells on ice and add an equal volume (~500 μl) of acid washed glass beads. Vortex the cells vigorously (in the cold room) for two cycles of 20 min with a 10 min break on ice. Be sure the beads don't settle during vortexing since a large amount of foaming occurs. Use ice cold glass beads to prevent heat formation during vortexing, which can cause physical distortion of chromatin structure.
Puncture the Eppendorf tube at the bottom using a heated 22 gauge needle and insert into a 15 ml centrifuge tube. Centrifuge in the Sorvall SH3000 at 3,000 × g for 10 min at 4°C. Collect the lysate in a 15 ml tube. Discard the Eppendorf tube containing the glass beads. Resuspend the loose pellet and transfer the entire lysate to an Eppendorf tube. Centrifuge at 13,000 g in the microfuge for 10 min at 4°C. Expect to see a thin transparent layer of chromatin on the top of a turbid pellet of cell debris.
Discard the supernatant and resuspend the pellet in 1 ml FA lysis buffer. Centrifuge at 13,000 g in the microfuge for 10 min at 4°C. Discard the supernatant, resuspend the pellet thoroughly in 500 μl 10 mM Tris·HCl (pH 7.5). Chromatin samples at this stage can be stored in 50 μl aliquots at −80°C for several months.
3.4. Restriction digestion of crosslinked chromatin
To 50 μl of the crosslinked chromatin sample, add 10 μl of 10X restriction enzyme buffer and 30 μl of ddH2O. Mix well by pipeting up and down. Add restriction enzyme, mix well, and incubate for 5 h at 37°C with gentle shaking. We often use 10 μl of EcoRI (20U/μl) or HindIII (20U/μl). Monitor the extent of restriction digestion by convergent PCR. The kinetics of chromatin digestion at the SAC3, BLM10, GAL10 and HEM3 genes, using either EcoRI or HindIII, are depicted in Fig. 2.
To detect looping at genes with short ORFs, it is critical to perform restriction digestions for 4–5 hr. Digestion of crosslinked chromatin for less than 4 hr is not adequate for detection of short-range interactions. Add 10 μl of 10% SDS and heat the sample at 65°C for 20 min to inactivate the restriction enzyme. Add 565 μl of ddH2O and 75 μl of 10% Triton X-100 to sequester the SDS. Centrifuge the sample at 13,000 g for 20 min at room temperature. Resuspend the pellet thoroughly in 100 μl 10 mM Tris·HCl (pH 7.5) by pipeting up and down.
Alternatively, the whole digested chromatin sample (without centrifugation) can be used in the subsequent ligation step. However, ligation in this solution occurs less efficiently than it does using the resuspended pellet. We have also performed 3C analysis using the supernatant fraction following centrifugation of digested chromatin. For example, in Fig. 6 of Ref. 21, we mapped loops at SEN1 using the supernatant fraction, whereas in Fig. 3. of Ref. 22, we mapped loops at the BLM10 and HEM3 genes using the resuspended pellet. Although the results are qualitatively similar among the different procedures, we find that the subsequent ligation reaction occurs most efficiently using the resuspended pellet, presumably because materials that interfere with ligation have been depleted from the pellet.
3.5. Intramolecular ligation of cross linked chromatin
To 100 μl of digested chromatin add 350 μl of Quick Ligase Buffer, 245 μl of ddH2O and 5 μl of Quick T4 DNA ligase (total volume = 700 μl) . Mix thoroughly and incubate the sample for 1 hr at 25°C. The ligation reaction must be done in dilute solution to minimize intermolecular ligation of non-crosslinked DNA. Quick DNA ligase gives better and more reproducible results than regular T4 ligase and improves the sensitivity of the 3C signal for detection of short range interactions.
3.6. Reversal of chromatin crosslinks
To ensure complete removal of RNA, add 2 μl of 10 mg/ml DNase-free RNase to the reaction mixture. Incubate at 37°C for 10 min. To reverse crosslinks, add 7 μl of 10% SDS and 5 μl of 10 mg/ml proteinase K to the ligation mixture, mix well and incubate overnight at 65°C in a water bath. Add an equal volume (700 μl) of phenol:chloroform:isoamyl alcohol (25:24:1) to the reaction mixture. Vortex the sample for 20 sec, and centrifuge at 10,000 g for 10 min at room temperature. Collect the upper aqueous phase. Be careful to avoid the interface layer as the aqueous phase will appear turbid. Repeat the phenol:chloroform:isoamyl alcohol extraction one time, followed by chloroform/isoamyl alcohol extraction one time to remove the residual phenol.
Transfer the aqueous phase to a fresh Eppendorf tube, add 1/10 volume of 3 M sodium acetate (pH 5.2) and 2 μl of glycogen (20 mg/ml). Vortex briefly. Add 2.5 volumes of ethanol and mix gently by inverting the tube. Incubate for 20 min at room temperature and centrifuge for 20 min at 13,000 g at room temperature. Precipitation of DNA below room temperature increases salt precipitation. It is therefore preferable to precipitate the DNA at room temperature. Ethanol precipitation of DNA at the final step should be performed in the presence of a carrier. Nuclease-free glycogen gives consistent results.
Decant the supernatant by gently inverting the tube to avoid dislocating the tiny pellet. Re-centrifuge at 13,000 g for 10 min at room temperature. Carefully withdraw the remaining supernatant using a 100 μl pipette. Air-dry the pellet completely and dissolve in 50 μl TE (pH 8.0). This is now the 3C DNA template, which can be stored at this stage at −20°C for several months.
Determine the DNA concentration by absorption spectroscopy at 260 nm. Typically 250–300 ng of the DNA template is required for 3C PCR reactions. Titration of the DNA template must be done to assure a linear relationship between the amount of DNA and the resulting PCR products. The same amount of template DNA should be used in all subsequent PCR reactions.
3.7. Locus-specific PCR
Thaw the reaction components on ice and vortex well before use. For each PCR reaction mix 5 μl 10X PCR buffer, 1 μl 10 mM dNTP mix, 1 μl of each primer pair (25 pmol), 0.5 μl of Taq DNA polymerase (5U/μl), 300 ng of 3C DNA template and ddH2O to bring the total volume to 50 μl. Tap to mix, then centrifuge briefly in a microcentrifuge to collect reagents at the bottom of the tube.
Program the thermal cycler for hot-start, typically 94°C for 2 min, followed by 30 cycles at 94°C for 30 sec, 52°C for 30 sec, 72°C for 30 sec, then final extension at 72°C for 5 min. PCR conditions can vary, of course, according to primer Tm and product length. When the PCR amplification is complete, add 5 μl of 6X gel loading buffer without bromophenol blue and briefly centrifuge to mix. Separate the PCR products in a 1.5% agarose gel containing 1X TBE and 0.5 μg/ml ethidium bromide, typically running the gel at 100 volts for 2 hr. Results from 3C analysis of juxtaposed regions of the BLM10 gene are shown in Fig. 3.
Quantify the PCR products using a gel documentation system containing appropriate software. The 3C interaction frequency for each primer pair is determined as the ratio of the 3C PCR product to the control PCR product (Fig. 3C and 3D). Setup the PCR reactions in triplicate to determine average interaction frequency. Accordingly, the relative abundance of the PCR products is proportional to the frequency with which the chromatin regions interact.
4. Concluding remarks
3C is a straightforward, powerful technique to determine the frequency of physical interaction between any two regions within the genome of any organism. We have adapted 3C to explore dynamic interactions at single gene loci in the yeast S. cerevisiae. Using 3C, we have detected gene loops at the SEN1 (6.7 kb), BLM10 (6.4 kb), BUD3 (4.9 kb), SAC3 (3.9 kb), GAL10 (2.1 kb) and HEM3 (1.0 kb) genes (21, 22). In each case, 3C analysis produced robust PCR signals using primers specific for distal ends of the genes. Using a common primer specific to either the promoter or terminator, paired with primers that “walk” across the gene (for example, Fig. 1, primer pairs b & e, b & f, b & g and b & d, or primer pairs c & h, c & i, c & j and c & a), we established that gene loops juxtapose specifically promoter and terminator regions of the SEN1, BLM10 and HEM3 genes (21, 22), as well as the BUD3 and MET16 genes (A. Ansari, unpublished results). 3C analysis from the Proudfoot and Mellor laboratories also demonstrated promoter-terminator gene looping at the FMP27 and SEN1 genes (27). Although we initially chose exceptionally long genes for 3C analysis in an effort to minimize crosslink-independent interactions, we are now able to detect promoter-terminator interactions for genes less than 1 kb in length.
We have successfully used 3C to identify factors involved in loop formation. Using the same target genes described above, we found that dynamic promoter-terminator interactions are diminished or eliminated in an rpb1–1 mutant that is defective for transcription, and in pta1 and ssu72 mutants that are defective for mRNA 3'-end processing (21, 22). Thus, gene loops are transcription-dependent and involve components of the termination machinery. Moreover, a mutant defective for the transcription initiation factor TFIIB is also impaired for looping (22). Taken together, these results suggest gene loops are dynamic structures whose formation involves physical interaction between the transcription initiation and termination machineries.
We suspect that gene loops are a common feature of genes transcribed by RNA polymerase II and not unique to yeast. The HIV-1 provirus was recently reported to form a loop structure between the 5' LTR promoter and 3' LTR poly(A) signal, also in a transcription-dependent manner (28). Dynamic promoter-terminator loops have also been demonstrated for the mammalian BRCA1 gene (29) and the gene encoding the CD68 immunohistological marker (30). These results suggest that gene loops might be a general feature of active gene expression.
Our 3C protocol should be amenable for use by any laboratory to investigate dynamic short-range chromatin interactions. Furthermore, we anticipate that our protocol can be adapted to study gene looping in metazoan organisms simply by defining the conditions for crosslinking and isolating chromatin, specific to each cell type. 3C can now be applied to identify gene loops at any locus, to determine the factors involved in looping, and to define the physiological conditions under which loops are formed and maintained.
Supplementary Material
Acknowledgments
We are grateful to Krishnamurthy Shankarling (RWJMS), Claire Moore (Tufts Medical School) and the members of our laboratories for many helpful discussions during the course of this work. This work was supported by NIH RO1 grants GM39484 (to M.H.) and GM068887 (to Claire Moore and M. H.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kornberg R. Angew Chem Int Ed Engl. 2007;46:6956–65. doi: 10.1002/anie.200701832. [DOI] [PubMed] [Google Scholar]
- 2.Spector DL. Annu Rev Biochem. 2003;72:573–608. doi: 10.1146/annurev.biochem.72.121801.161724. [DOI] [PubMed] [Google Scholar]
- 3.Chambeyron S, Bickmore WA. Curr Opin Cell Biol. 2004;16:256–62. doi: 10.1016/j.ceb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Fraser P. Curr Opin Genet Dev. 2006;16:490–5. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Misteli T. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 6.Marenduzzo D, Faro-Trindade I, Cook PR. Trends Genet. 2007;23:126–33. doi: 10.1016/j.tig.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Dekker J. Science. 2008;319:1793–4. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dekker J, Rippe K, Dekker M, Kleckner N. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 9.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nat Genet. 2006;38:1348–54. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Nat Genet. 2006;38:1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 11.Dostie J, Dekker J. Nat Protoc. 2007;2:988–1002. doi: 10.1038/nprot.2007.116. [DOI] [PubMed] [Google Scholar]
- 12.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Genome Res. 2006;16:1299–309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dostie J, Zhan Y, Dekker J. Curr Protoc Mol Biol. 2007 doi: 10.1002/0471142727.mb2114s80. Chapter 21, Unit 21 14. [DOI] [PubMed] [Google Scholar]
- 14.Gondor A, Rougier C, Ohlsson R. Nat Protoc. 2008;3:303–13. doi: 10.1038/nprot.2007.540. [DOI] [PubMed] [Google Scholar]
- 15.Tiwari VK, Cope L, McGarvey KM, Ohm JE, Baylin SB. Genome Res. 2008;18:1171–9. doi: 10.1101/gr.073452.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Splinter E, Grosveld F, de Laat W. Methods Enzymol. 2004;375:493–507. doi: 10.1016/s0076-6879(03)75030-7. [DOI] [PubMed] [Google Scholar]
- 17.Simonis M, Kooren J, de Laat W. Nat Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 18.Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. Nat Protoc. 2007;2:1722–33. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 19.Dekker J. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 20.Miele A, Gheldof N, Tabuchi TM, Dostie J, Dekker J. Curr Protoc Mol Biol. 2006 doi: 10.1002/0471142727.mb2111s74. Chapter 21, Unit 21 11. [DOI] [PubMed] [Google Scholar]
- 21.Ansari A, Hampsey M. Genes Dev. 2005;19:2969–78. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh BN, Hampsey M. Mol Cell. 2007;27:806–16. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Winston F, Dollard C, Ricuperohovasse SL. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 24.Thomas BJ, Rothstein R. Cell. 1989;56:619–30. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 25.Solomon MJ, Varshavsky A. Proc Natl Acad Sci U S A. 1985;82:6470–4. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman F. Methods. Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 27.O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Nat Genet. 2004;36:1014–8. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 28.Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. Mol Cell. 2008;29:56–68. doi: 10.1016/j.molcel.2007.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Proc Natl Acad Sci U S A. 2008;105:5160–5. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Reilly D, Greaves DR. Genomics. 2007;90:407–15. doi: 10.1016/j.ygeno.2007.04.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.