Fig. 1.
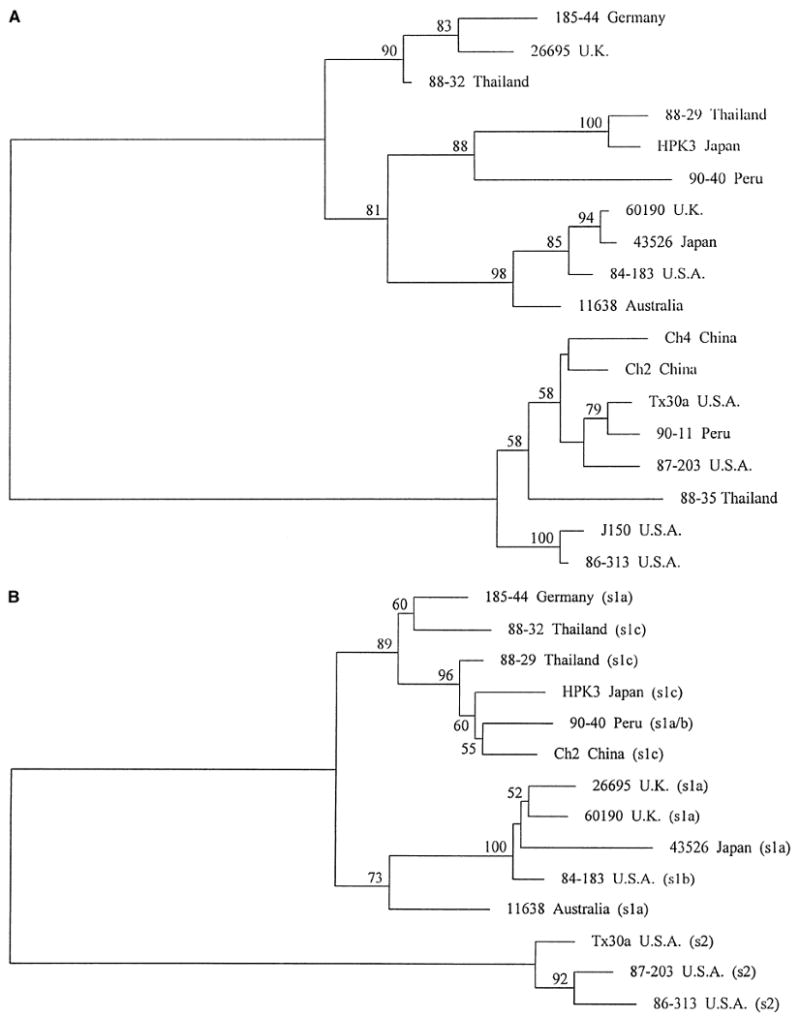
Phylogenetic relationships among strains based on two adjacent sections of the vacA mid region sequence. Trees were mid-pointed rooted, separating the vacA type m1 strains (upper cluster) from the vacA type m2 strains (lower cluster). Horizontal distances between strains indicate relatedness. Numbers on branches indicate the percentage of bootstrap replicates in which the clusters to the right were found (only values greater than 50% are shown); in both cases, the separation of the two main clusters occurred in 100% of bootstraps. Panel A is based on multiple sequence alignments of part of the distal mid region of vacA (region A, coordinates 1971–2259 bp in strain Tx30a [1]). Panel B is based on the part of the vacA mid region immediately 5′ region to that analyzed in Panel A (region B, coordinates 1606–1970 bp in strain Tx30a [1]). Five m2 strains are not included in Panel B because sequence data were not obtained for them for this region. The type of signal sequence coding region in each strain, as determined by nucleotide sequence analysis, is added in brackets. Strain 90-40 has a signal region type intermediate between s1a and s1b.
