Fig. 3.
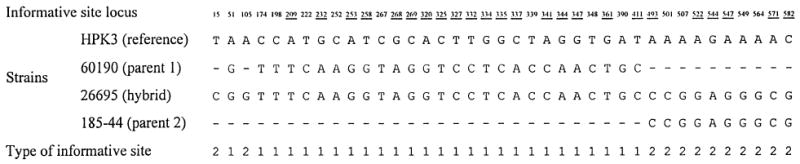
Informative site analysis to demonstrate the hybrid nature of vacA in strain 26695, the strain for which the complete genome sequence has been determined [25]. Dashes represent nucleotide identity with the reference sequence (HPK3). Strains 60190 and 185-44, which cluster most closely with 26695 in Fig. 1B and 1A, respectively, were taken as parent-like strains, and strain HPK3, which clusters distantly in both trees, was used as a reference strain. Only informative sites (sites where two pairs of strains have identical nucleotides) are shown, and these are numbered from the beginning of the concatenated sequence of regions A and B analyzed in Fig. 1 (coordinates 1606–2259 bp in strain Tx30a [1]). Type 1 sites suggest clustering of 26695 with 60190, type 2 with 185-44, and type 3 (of which there are none) with HPK3. Underlined and emboldened loci are non-synonymous substitutions compared with the other pair of sequences. The distribution of type 1 and 2 informative sites is clearly non-random (p < 10−4) and shows a recombination point somewhere between sites 411 and 493 of the concatenated sequence.
