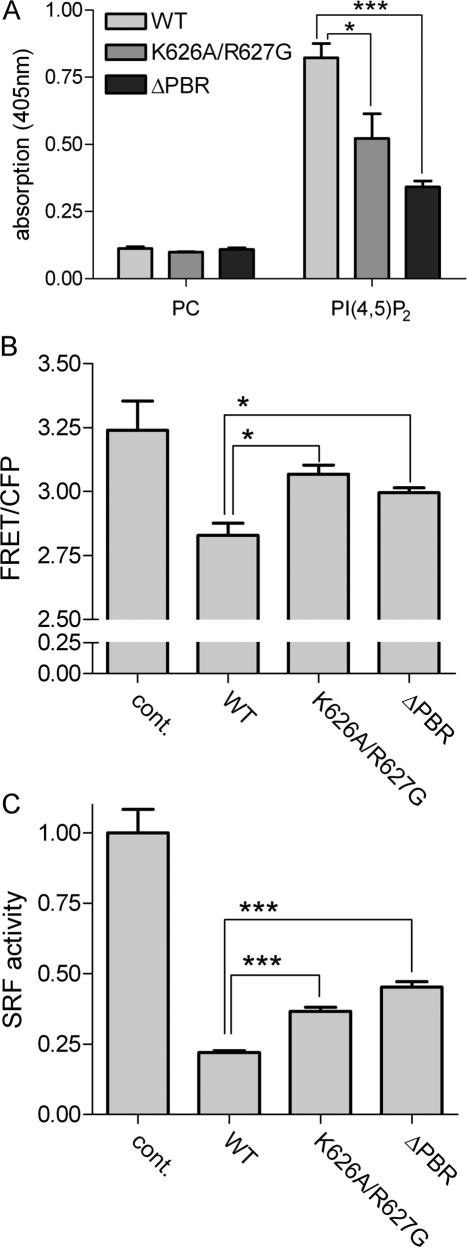
Figure 5.
The DLC1 PBR is required for RhoGAP activity in intact cells. (A) GFP-tagged DLC1 WT, K626A/R627G, and ΔPBR were tested for their ability to bind immobilized PI(4,5)P2 as described in Figure 1. (B) HEK293T cells were cotransfected with plasmids encoding the Raichu-RhoA biosensor and Flag-DLC1 WT, K626A/R627G, ΔPBR, or empty vector (cont.). The next day, the emission ratio of Raichu-RhoA was determined by measuring YFP (FRET) and CFP fluorescence (excitation 433 nm) in cell lysates. One representative experiment out of three is shown; error bars, SEM. (C) HEK293T cells were transiently transfected with the 3DA.Luc reporter, a plasmid encoding for Renilla luciferase and empty vector (cont.), pEGFPC1 DLC1 WT, K626A/R627G, or ΔPBR, respectively. Cells were serum-starved overnight and then restimulated with 15% serum for 6 h. Firefly luciferase activity was determined and normalized by Renilla luciferase activity. The control was set to 1. Equal expression of the DLC1 variants was verified by measuring GFP fluorescence in the lysates with a fluorescence spectrometer. One representative experiment out of three is shown, each performed with triplicate samples; error bars, SEM.