Figure 2.
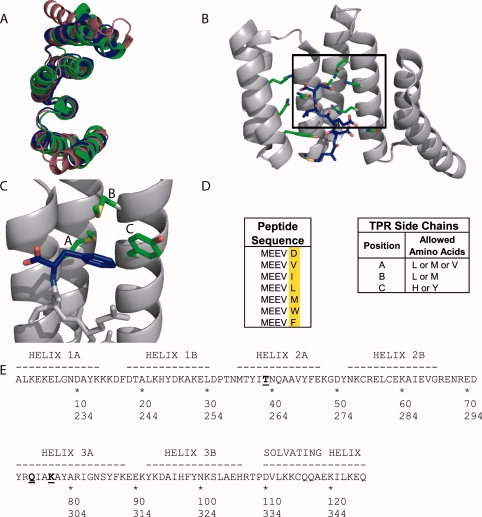
The TPR Scaffold and Library Design. (A) Overlay of the crystal structures of three natural 3-TPR domains: from PP5 (magenta, PDB file 1A17)6, TPR1 domain of HOP (blue, PDB file 1ELW)3, and TPR2A domain of HOP (green, PDB file 1ELR)3. The TPR1 and TPR2A structures are taken from the structures of the TPR in complex with its peptide ligand. The PP5 structure is of the TPR domain alone. (B) The cocrystal structure of TPR2A (gray ribbon) with its cognate ligand, the C-terminal peptide of Hsp90 (MEEVD) (blue sticks).3 Some of the side chains of TPR2A involved in interactions with the peptide, including all residues varied in this work, are shown as sticks. (C) Illustration of the hydrophobic cavity of a designed TPR in complex with the MEEVF peptide. This figure should not be interpreted as an optimal model for the T-Mod-peptide interaction, but instead a guide for the reader. The randomized TPR positions are shown in green and labeled A-C, and the C-terminal residue of the peptide is shown in blue with the carboxylate oxygen atoms in red. Images A-C were rendered in PyMol. (D) Sequences of peptides (left) and TPRs (right) used in this study. A library of 12 TPR proteins was constructed based on the sequence of TPR2A with randomizations introduced to the hydrophobic residues listed. (E) The sequence of TPRs used in this study compared to wild-type TPR2A.3 Wild-type TPR2A was used as the scaffold, with a hydrophobic cavity introduced by randomizing three key positions shown in bold and underlined. In addition, the mutation D110K (equivalent to position 334 in HOP) was introduced which gives a slight increase in the affinity of wt-TPR2A for Hsp90 (Tommi Kajander and Lynne Regan, manuscript in preparation). The first line gives the numbering scheme based on TPR2A alone, the second line shows the corresponding numbering in the full-length HOP protein. TPRs are labeled in the text based on the residues at the three randomized positions in the format (T-Mod(ABC)).
