Table I.
Data Collection and Refinement Statistics
| Data Collection Statistics | |
| Space group | P21 |
| Unit cell | a = 37.6 Å, b = 66.6 Å, c = 48.6 Å, β = 107.21° |
| Wavelength (Å) | 1.54178 |
| Resolution (Å) | 50–2.2 (2.8–2.2) |
| Signal/noise (1/σ) | 25.8 (4.9) |
| Completeness (%) | 98.6 (87.4) |
| Rsyma (%) | 5.3 (18.7) |
| Redundancy | 5.7 |
| Unique reflections | 11,529 |
| Refinement statistics | |
| Number of molecules per asymmetric unit | 2 |
| Number of atoms | |
| Total | 2332 |
| Protein | 2195 |
| Peptide | 48 |
| Waters | 85 |
| Ni | 4 |
| RMSD bond lengths | |
| Bond lengths (Å) | 0.0009 |
| Bond angles (°) | 1.2 |
| Average B value | 14.8 |
| RWorking (%) | 18.4 |
| RFreeb (%) | 24.4 |
| Ramachandran plot (2 complexes per asymmetric unit) | |
| Residues in the most favored regions | 255/260 |
| Residues in the additional allowed regions | 5/260 |
| Residues in the generously allowed regions | 0 |
| Residues in the disallowed regions | 0 |
The values in parenthesis are for the highest resolution bins.
Rsym = 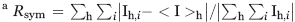 , where <I>h is the average intensity of symmetry-related reflections.
, where <I>h is the average intensity of symmetry-related reflections.
Rfree was calculated the same as Rwork but on 5% of randomly selected data not used in refinement.
