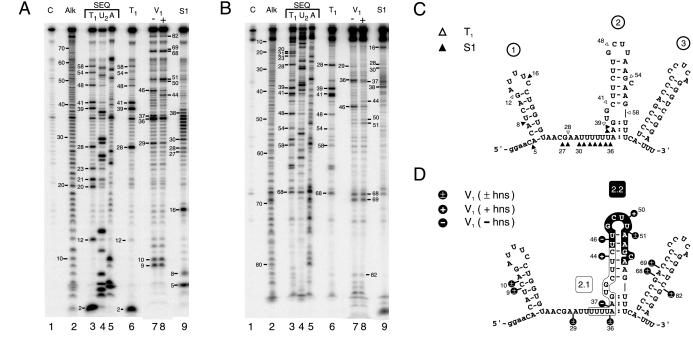
Figure 3.
Nuclease footprinting of DsrA. DsrA substrate RNA produced and end-trimmed in vitro was labeled at the 5′ or 3′ end, then treated with ribonucleases. Digestion products are numbered by phosphodiester bond position from the 5′ end. (A) Footprinting of 5′-labeled DsrA. (B) Footprinting of 3′-labeled DsrA. Lane numbers are given below, with reactions specified above. Lane C, untreated control RNA; Alk, alkaline hydrolysis ladder; lanes 3–5, sequencing reactions performed under denaturing conditions; and lanes 6–9, specific nuclease footprinting reactions performed under nondenaturing conditions. Lanes 3 and 6, RNase T1; lane 4, RNase U2; and lane 5, RNase A; lanes 7 and 8, double-strand-specific RNase V1 reactions performed in the absence or presence, respectively, of unlabeled hns RNA; and lane 9, S1 nuclease. (C and D) Predicted secondary structure of DsrA based on single-strand- and double-strand-specific nuclease susceptibility. (C) Single-strand-specific nuclease susceptibility. ▵, RNase T1; and ▴, S1 nuclease. (D) Double-strand-specific nuclease susceptibility. The hns-complementary region characterized (4) is shown boxed (region 2.1) whereas the newly proposed hns-complementary sequence (region 2.2) is shown in white letters on a black background. Cleavage sites are indicated by black lollipops. +, Cleavage only in the presence of hns RNA; −, cleavage only in the absence of hns RNA; ±, cleavage occurs in the presence or absence of hns RNA; and :, partial double-strand character.