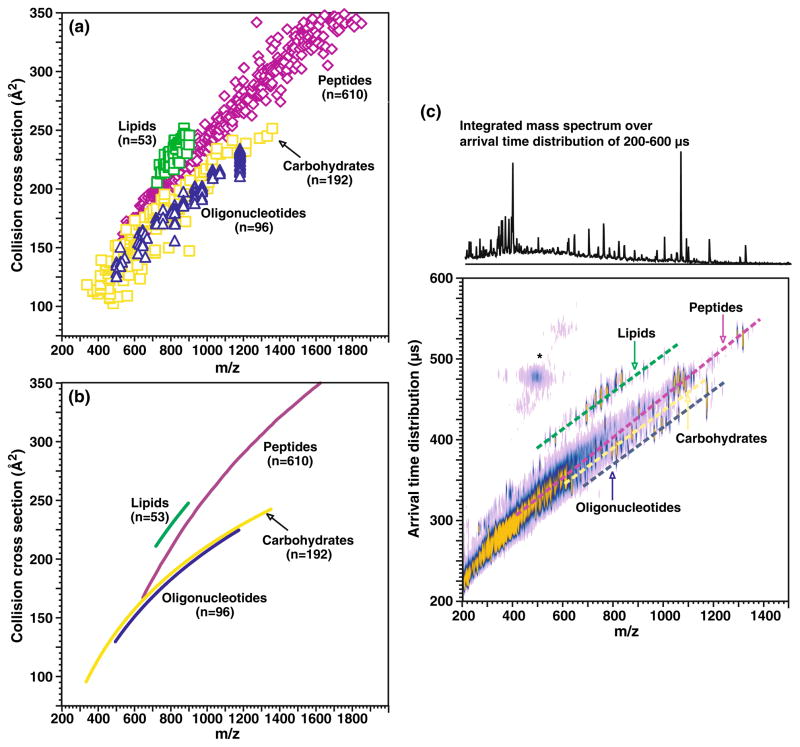
Fig. 1.
a A plot of collision cross section as a function of m/z for different biologically relevant molecular classes, including oligonucleotides (n=96), carbohydrates (n=192), peptides (n=610), and lipids (n=53). All species correspond to singly charged ions generated by MALDI, where error ±1σ is generally within the data point. Values for peptides species are obtained from a previous study (see Electronic Supplementary Information Table S1 for numerical data) [68]. b A plot of the average collision cross section versus m/z fitted to logarithmic regressions for the data corresponding to each molecular class. The specific equations for each class are as follows: oligonucleotides y=107.45 ln(x)−535.34 (R2=0.96), peptides y=197.4 ln(x)−1,109.8 (R2=0.96), carbohydrates y=103.7 ln(x)−507.22 (R2=0.83), and lipids y=164.59 ln(x)−871.44 (R2=0.70), respectively. c A plot of MALDI-IM-MS conformation space obtained for a mixture of model species representing each molecular class (ranging from seven to 17 model species for each class, spanning a range of masses up to 1,500 Da). Dashed lines are for visualization purposes of where each molecular class occurs in conformation space. Signals in the vicinity of the asterisk arise from limited post-IM fragmentation of the parent ion species