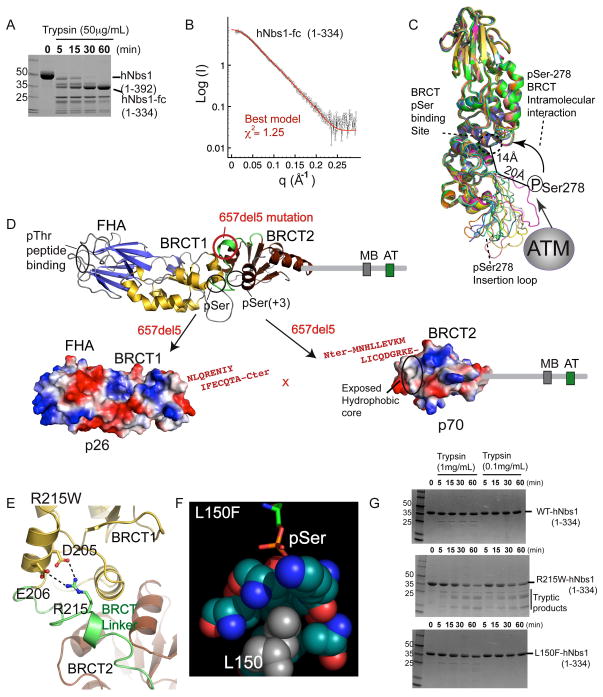
Figure 2. Structural Basis for NBS Mutations.
(A) Trypsin degradation produces a stable hNbs1 folded core (hNbs1-fc, residues 1-334).
(B) Comparison of calculated SAXS curves for hNbs1 models (green and red) with experimental scattering (black) shows hNbs1-fc architecture closely matches the S. pombe structure.
(C) Superposition of 12 homology hNbs1 models with SAXS scattering χ2 fits better than 1.3. ATM Phosphorylation sites in hNbs1 map to surface accessible loops with observed conformational variability between models. Ser278 phosphorylation could mediate auto-regulatory BRCT peptide binding in cis through an interaction with the phosphoprotein-binding cleft.
(D) Nbs1 657del5 frameshift mutation found in 90% of NBS patients generates protein fragments p26 and p70. Sequence in red, resulting from 657del5, shows extra C-terminal residues added to the N-terminal (p26) and C-terminal (p70) fragments.
(E) A homology model for Arg215 in hNbs1 based on the spNbs1-fc structure suggests Arg215 forms a salt bridge from the BRCT linker to a BRCT1 helix. R215W is predicted to disrupt protein folding.
(F) The Nbs1 L150F missense mutation likely disrupts the BRCT pSer binding pocket. Leu150 is found in the hydrophobic core directly contacting pSer ligands, based on the structure of a BRCA1-phosphopeptide complex, RCSB ID 1T2V.
(G) Protease sensitivity of Nbs1 missense mutations. Mutant R215W destabilizes the protein fold whereas L150F shows WT resistance to trypsin digestion.