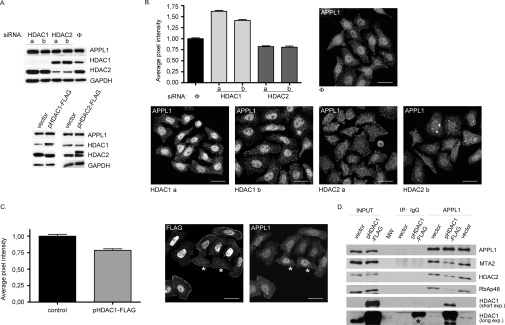
Figure 3. The interactions with NuRD affect cellular distribution of APPL1.
(A) APPL1 protein levels do not depend on HDAC1 or HDAC2. Extracts of HeLa cells with reduced HDAC1 or HDAC2 levels by siRNA [two different oligonucleotides (a, b) per gene or non-specific oligo (Φ), transfected for 72 h; top panel] and HEK-293 cells transfected for 48 h with plasmids encoding FLAG-tagged HDAC1 (pHDAC1–FLAG), HDAC2 (pHDAC2–FLAG) or with a control vector (bottom panel), were analysed by Western blotting using anti-APPL1 antibodies. To demonstrate the efficiency of silencing or overexpression, extracts were probed using anti-HDAC1 and anti-HDAC2 antibodies. GAPDH was included as a loading control. (B) Microscopy-based analysis of APPL1 nuclear localization upon silencing of HDAC1 or HDAC2. HeLa cells were transfected with siRNA oligonucleotides: one non-specific (Φ) and two specific per gene (a and b) against HDAC1 and HDAC2 for 72 h, followed by fixation and immunostaining for APPL1. Acquired microscopic images were analysed by Metamorph software and the average pixel intensities corresponding to APPL1 in the nuclei (as visualized by Hoechst staining, not shown) were calculated. The results of a representative experiment are shown in the graph. The values are normalized with respect to the average pixel intensity of nuclear APPL1 in cells transfected with non-specific siRNA, assigned one arbitrary unit. Error bars indicate standard error (minimum 100 cells from each transfection were used for the analysis). The results were statistically analysed by GraphPad Prism4 software, and the values obtained for each knockdown experiment were significantly different from the control at P<0.0001. The images demonstrate the cellular localization of APPL1 upon silencing of HDAC1 or HDAC2 (as indicated) and represent a maximal projection of z-stacks. Scale bar: 24 μm. (C) HeLa cells transfected with the plasmid encoding FLAG-tagged HDAC1 (pHDAC1–FLAG) were analysed with respect to the nuclear localization of APPL1 as described above. Untransfected cells (marked with asterisks) were used as a control (the average pixel intensity of nuclear APPL1 set to one unit). The statistical analysis was performed on 50 transfected and 50 untransfected cells, and the difference between them was statistically significant at P<0.0001. The images represent a maximal projection of z-stacks. FLAG staining (left image) is shown to discriminate between transfected and untransfected cells with respect to APPL1 staining (right image). Scale bar: 24 μm. (D) HDAC1 overexpression leads to the destabilization of binding between APPL1 and HDAC2-containing NuRD complex. Extracts of HEK-293 cells transfected with HDAC1-FLAG or with a control vector were subjected to immunoprecipitation (IP) with anti-APPL1 or non-specific (IgG) antibodies. Immunoprecipitates, along with 10% of the extracts used (input), were analysed by Western blotting using the indicated antibodies. MW indicates a lane loaded with a molecular mass (weight) marker. Two exposures (short and long) of the HDAC1 blot are shown. Overexpressed HDAC1 exhibits some non-specific binding to IgG-covered protein G beads (visible at the long exposure of the blot and marked with an asterisk); however, its binding to beads containing anti-APPL1 antibodies is higher.