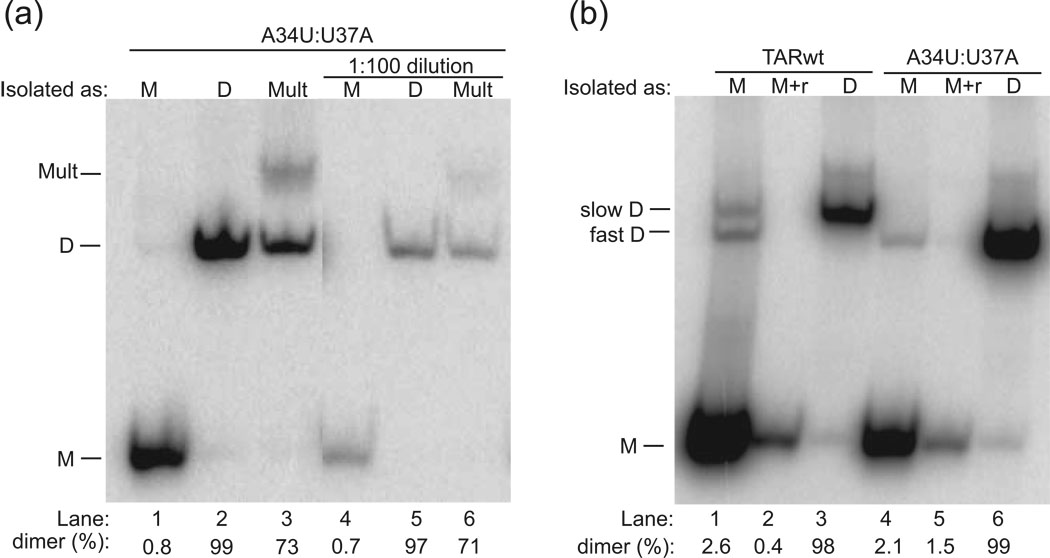
Fig. 3.
Analysis of native gel-purified RNAs. The form in which the band was isolated is provided following ‘Isolated as:’. (a) Analysis of native gel-purified A34U:U37A. Purification as well as dilution does not affect the integrity of A34U:U37A monomer (M) (lanes 1 and 4) or dimer (D) (lanes 2 and 5), but the multimer (Mult) is converted mostly to dimer (lanes 3 and 6). (b) Analysis of native gel-purified TARwt and A34U:U37A. Occasionally, bands isolated as monomer contained one or more trace dimer bands when re-fractionated (Lanes 1 and 4), but this could be removed by further renaturing these samples (incubating at 90 °C for 3 min/ room temperature 10 min) in 1×TE at concentrations of 1–3 µM (denoted ‘M+r’) (Lanes 2 and 5). In addition, migration of TARwt dimer is slower than A34U:U37A dimer (Lanes 3 and 6). Analysis of native-gel-purified single mutants is provided in the Supplementary Fig. 1. All gels are 10% polyacrylamide native gels (0.5×TBE) fractionated for ~2 h at 16 °C. The percent of strands in dimer is provided below the gel.