Abstract
Residual dipolar couplings (RDCs) between NC’ and NCα atoms in polypeptide backbones of proteins contain information on the orientation of bond vectors that is complementary to that contained in NH RDCs. The 1JNCα and 2JNCα scalar couplings between these atoms also display a Karplus relation with the backbone torsion angles and report on secondary structure. However, these N-C couplings tend to be small and they are frequently unresolvable in frequency domain spectra having the broad lines characteristic of large proteins. Here a TROSY-based J-modulated approach for the measurement of small 15N-13C couplings in large proteins is described. The cross-correlation interference effects inherent in TROSY methods improve resolution and signal to noise ratios for large proteins, and the use of J-modulation to encode couplings eliminates the need to remove frequency distortions from overlapping peaks during data analysis. The utility of the method is demonstrated by measurement of 1JNC’, 1JNCα, and 2JNCα scalar couplings and 1DNC’ and 1DNCα residual dipolar couplings for the myristoylated yeast ARF1·GTPγs protein bound to small lipid bicelles, a system with an effective molecule weight of ~70 kDa.
Keywords: RDC, Bicelle, myristoylated protein, scalar coupling, J-modulation, TROSY
1. Introduction
Couplings between pairs of spin ½ nuclei are a rich source of information for the structural characterization of proteins. The most common applications involve the use of three bond scalar couplings to restrict torsional angles about the central bond, such as the use of 3J between an amide proton and a Hα proton to restrict the ϕ angle between Cα and its directly bonded nitrogen [1,2,3,4], and the use of residual dipolar couplings (RDCs) to restrict orientations of bond vectors between pairs of spin ½ nuclei, such as that between an amide proton and an amide nitrogen of a 15N labeled protein [5,6,7,8]. Interestingly the 1JNCα and 2JNCα scalar couplings are similar to three bond scalar couplings in their dependence on backbone dihedral angles, but depend on Ψi and Ψi−1 respectively, instead of ϕ [9,10,11]. Likewise, 15N-13C RDCs provide complementary information to 1H-15N and other larger RDCs in restricting orientations of backbone structural elements [8,12,13]. The heteronuclear dipolar coupling Hamiltonian and the weak J-coupling Hamiltonian actually have the same spin part, so measurements of RDCs and J couplings usually utilize the same NMR experiments. While most experiments work well for measurement of the larger couplings, application becomes challenging when couplings are small compared to the spectral line widths. This is particularly an issue for 15N-13C couplings in larger proteins. Here we present a set of experiments that prove to be of particular value in these situations.
There has, of course, been considerable work devoted to the design of experiments for the measurement of scalar and dipolar couplings. In the most straightforward applications the couplings are measured from the frequency separations of lines in multiplet structures. The amide 15N-1H J coupling of ~93Hz is usually larger than the 15N/1H line-widths of proteins studied by solution NMR and can be determined at relatively high accuracy. The 15N-1H RDCs are manifested as small changes in the doublet splitting and the slight increase in line-width in weakly orientated proteins do not severely degrade the measurement accuracy. The large 15N-1H J coupling relative to the 15N transverse relaxation also makes possible the use of an IPAP type experiment, in which the doublet components are separated into different spectra in order to minimize spectral overlap without significant loss of sensitivity [14]. In contrast, the N-C couplings (1JNC’ ≈ 15Hz, 1JNCα ≈ 11Hz, 2JNCα ≈ 7Hz) are small and measurement by frequency separation is usually limited to small proteins where sharp 15N lines are obtainable. The situation can be improved somewhat by allowing the J coupling to evolve longer than the chemical shift, thus scaling up the apparent splittings [15,16,17]. This does not solve the problem of doublet overlap because the line-width is often scaled up proportionally, however, the digitization error is reduced. IPAP type experiments can be used to eliminate overlap of peaks in 1JNC’ and 2JNCα doublets [11] but IPAP is not suitable for the measurement of 1JNCα since there is no effective way of selectively creating anti-phase magnetization for N-Cα(i) without also creating anti-phase magnetization for N-Cα(i-1). The HNCO-based E.COSY type experiments can also be utilized to separate the 2JNCα doublets through an extra C’ dimension subject to a much larger 1JC’Cα coupling[17]. However, due to the absence of a significant C’(i-1)-Cα(i) coupling, the E.COSY principle is inapplicable to the effective separation of the 1JNCα doublets and therefore its measurement is still limited by the 15N linewidth. Another complication associated with small J-couplings arises when the RDCs between pairs of atoms become comparable in size to their J couplings; this makes RDC sign and size determination ambiguous. In this case, E.COSY style experiments [18] can be employed if one of the weakly coupled spins is coupled to a third spin with a constant of known sign. The sign and size of an RDC can also be determined through variable angle sample spinning that scales anisotropic couplings but keeps scalar couplings unchanged [19].
As an alternative to frequency-based methods, J couplings can be obtained by intensity-based methods, including quantitative J-correlation [4,22] and J-modulation [10,20,21,24] experiments. The main difference between the two is that in the J-correlation method, the coupling is obtained from the ratio of two distinct functions correlated by J-coupling after a fixed through-coupling transfer period, while in the J-modulation method, only a single J-modulated function is recorded but at multiple transfer times. Because of its simplicity, the J-correlation method is widely used in 3-D based experiments. J-modulation is more often used in 2-D based experiments where a large number of modulation time points can be taken within a reasonable amount of spectrometer time. Because the coupling of interest is usually over-determined, the J-modulation experiment offers higher precision [21]. It also has the advantage that errors in derived couplings can be easily estimated based on the quality of multiple-point fitting. The intensity-based methods are applicable as long as chemical shift differences are larger than line-widths, regardless of the size of couplings. Thus they are widely used for the measurement of small homo- and heteronuclear couplings [10,20,22,23]. However, since longer modulation delays are necessary in order to measure small couplings, sensitivity loss from transverse relaxation degrades the performance. This is particularly a problem for large proteins and actually produces a similar problem to that encountered in frequency domain experiments when line widths approach the size of couplings to be measured.
The destructive interference between 1H-15N dipolar interaction and 15N CSA can significantly improve spectral resolution and enhance sensitivity for experiments involving long 15N transverse relaxation periods [25,26]. However, it is difficult to exclusively utilize the 15N TROSY component during the measurement of NH couplings. For example, in frequency separated experiments, at least one of the lines in the multiplet structure is contaminated by the fast anti-TROSY relaxation [14,27,28], while in J-modulated experiments, it is hard to avoid a proton π pulse that exchanges TROSY and anti-TROSY magnetizations [21]. In contrast, for the measurement of N-C couplings, the slow 15N TROSY relaxation can be utilized during the entire J-modulation delay; this allows the use of the long delay times needed for accurate measurement.
The experimental scheme presented here is directed at the measurement of small 15N-13C couplings in large proteins. The sequence is basically a constant/semi-constant time 1H-15N TROSY experiment [29] with an 15N evolution period modified to allow modulation by selected 13C couplings for various periods of time. The sensitivity is further enhanced by sharing 15N chemical shift evolution with the J-modulation periods. For the initial time points where the J-modulation periods are shorter than the time required for 15N chemical shift evolution, a semi-constant time evolution is automatically adapted. We also choose to collect a reference spectrum for every modulation delay, in which J couplings are refocused so that spectral intensity is only modulated by relaxation. The peak intensity ratio between the J-modulation spectrum and the reference spectrum eliminates the contribution from relaxation. The latter approach introduces characteristics of both J-correlated and J-modulated methods in that a reference function is employed to cancel the relaxation effects and multiple delays are used for non-linear fitting. In addition it eliminates complications due to line widths varying with J-modulation periods in the semi-constant time version, it improves fitting accuracy by removing the relaxation rate as a floating parameter, and it allows more flexibility in combining data from different experiments as may occur with unstable samples.
The procedure is illustrated with data on a GDP/GTP switch protein that is involved in vesicle trafficking, namely yARF1·GTP [29]. ARF1 is a 21 kDa member of the RAS super family. It is N-terminal myristoylated, and exposure of this myristoyl chain, in addition to an N-terminal amphipathic helix, is believed to modulate membrane association. We recently reported the solution structure for the GDP bound form of myristoylated yARF1 [31]. Here a bicelle associated GTPγs bound form of the myristoylated protein is used. The effective molecular weight of the complex is estimated to be 70 kDa, a large system that poses a good test for the pulse sequence.
2. Pulse Sequence Design
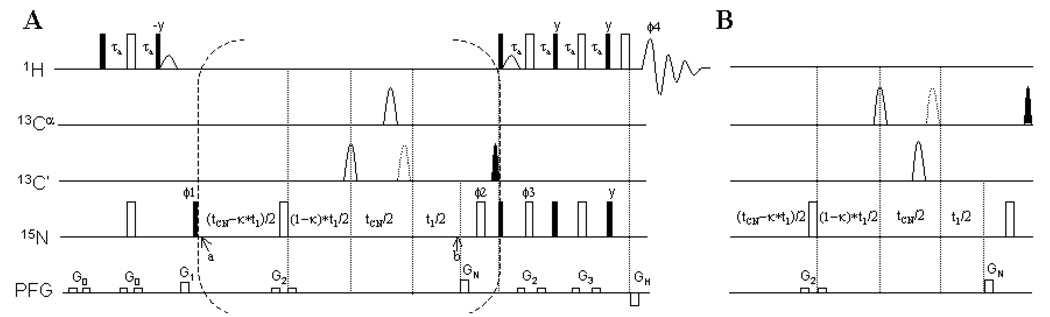
Figure 1A shows the pulse sequence for measurement of 1JNC’ coupling. The first four pulses on protons and two on nitrogen encompass a period in which an INEPT transfer of proton magnetization to nitrogen occurs. Between points a and b, within the brackets, 15N chemical shift evolves for t1, and 13C J coupling evolves for tCN in the coupling modulation experiment. In the 13C decoupled (reference) experiment the central 180° pulse is moved to the end of the tCN period which leads to complete refocusing of N-C couplings during the entire period when 15N magnetization is transverse, and a reference spectrum having only transverse relaxation but no J-modulation during tCN results. The 90° 13C pulse after point b creates 1HN-13C multiple quantum coherence from 1HN-13C coherence formed during the J-modulated experiment to ensure removal of artifacts from these components. The 15N in-phase (Nx/y ) to anti-phase (2Nx/yCz) transfer through the relaxation interference between 15N-13C dipole-dipole interaction and 15N CSA is negligible, so no attempt was made to remove these contributions. Since C’ and Cα carbons have no directly bonded protons under perdeuteration conditions, their longitudinal relaxation rates are small enough to satisfy that , so 15N transverse relaxation can be treated as the average of in-phase Nx/y and anti-phase 2Nx/yCz relaxation rates during modulation delays for both J-modulated and reference spectra. The pulses following the bracketed segment, coupled with the phase cycling described in the figure legend accomplish quadrature detection and selection of the TROSY component of the 1H-15N HSQC spectrum as described in the literature [29].
Figure 1.
Pulse sequences for the measurement of 1JNC’, 1JNCα, and 2JNCα couplings. A) the sequence for measurement of 1JNC’. B) the sequence to replace the part inside the dashed bracket of (A) to allow simultaneous measurement of 1JNCα, and 2JNCα couplings. Narrow solid and wide open bars represent 90° and 180° rf pulses, respectively. The pulses are x phased unless otherwise indicated. The scale factor κ is set to 1 if tCN is longer than the maximal t1 acquisition time t1,max, otherwise it is set to tCN/t1,max . The C’ 180° and 90° sinc pulses are centered at 174ppm with a null point at 56ppm. The Cα 180° and 90° reburp pulses are centered at 56ppm covering a band width of 50ppm. The sinc water flip-back pulse has a duration of 946µs. The delay τa is 0.91/(4JNH) ≈ 2.45 ms. The phase cycling is: Φ1 = {x, -x}, Φ2 = {2(x), 2(−x)}, Φ3 = {x} and Φ4 = {−x, x}. The gradient GH and the phase of Φ3 are inverted along with the echo/anti-echo acquisition. The z gradients are: G0 = 1.8G/cm, 0.5ms; G1 = 26.6G/cm, 1ms; G2 = 3.6G/cm, 0.5ms; G3 = 5.4G/cm, 0.5ms; GN = 31.9G/cm, 2ms; GH = 32.3G/cm, 0.2ms.
Simply swapping the selective pulses on C’ and Cα will allow simultaneous measurement of 1JNCα and 2JNCα couplings (Figure 1B) instead of 1JNC couplings. In principle, it is trivial to make a pulse sequence based on the current scheme for simultaneous measurement of 1JNC’, 1JNCα, and 2JNCα, e.g., by simultaneously inverting or exciting both C’ and Cα. However fitting coupling constants to modulation curves with a more complicated relationship involves additional parameters and could be less accurate.
Under the circumstances described above, the intensity ratio of the 13C J-modulated spectrum over the reference spectrum is:
| (1) |
where the expressions on the top and bottom describe respectively the intensity ratios from the 1JNC’ and 1JNCα-2JNCα experiments. The factor r takes into account non-ideal 13C π pulses and incomplete 13C enrichment. It varies between 0 and 1, with 1 corresponding to an ideal case in which the efficiency of both 13C incorporation and π pulses is 100%. The details are discussed in Appendix A. To ensure that r always falls in the legal range between 0 and 1, the following equations, in which cos2(a) is substituted for r, were used during the actual fitting process:
| (2) |
Obviously, Eq.(1) and Eq.(2) describe the same relationship except that the coefficients in Eq.(2) are automatically confined within the correct range. The intensity ratio between the J-modulated and reference experiments depends on some fixed experimental parameters, namely the 13C percentage, 13C π pulse efficiency and J couplings of interest, but is indifferent to conditions that can affect the absolute signal intensities or relaxation properties. As a result, experiments involving samples of different concentrations can be combined. Experiments collected at different B0 fields may also be combined, provided the 13C inversion pulses are properly calibrated to ensure nearly identical inversion efficiency. The couplings between amide 15N and Hα/Hβ in cases of incompletely perdeuterated or nondeuterated proteins are active only for a duration of t1 due to the presence of an 15N π pulse inside the J-modulation delay. Thus these couplings do not cause intensity modulation but merely some line-broadening in the 15N dimension, which does not affect the J-modulated measurement of NC couplings.
Residual dipolar couplings (RDCs) share spin operators with the Hamiltonian for first order scalar coupling and simply add to the observed splittings from scalar couplings. Hence, equation 1 and equation 2 apply equally well to cases where RDCs contribute with J+D replacing expressions in J alone.
3. Experimental
3.1 Sample Preparation
A His-tagged version of yARF1 was expressed in a deuterated, 15N, 13C enriched medium as previously described [31]. It was purified on a Hisx6 column and the His-tag removed by treatment with thrombin. The protein was further purified with a Q-sepharose column. The purified yARF1 which was pre-loaded primarily with GDP was then converted to a GTPγs form, that is only soluble in the presence of lipids, using the following procedure: A lipid mixture of DMPC/DHPC was added from a 40% (w/v) stock ([DMPC]:[DHPC] (q) =0.25) to a solution of the isolated yARF1. The resulting sample contains ~0.8mM yARF1·GDP, 10% (w/v) DMPC/DHPC (q=0.25), 10mM K2HPO4-KH2PO4 (pH 7), 50mM NaCl, 10mM K2SO4, 2mM MgCl2, 5% D2O, and 5mM dithiothreitol. The slowly hydrolyzing GTP analog, GTPγs, was then added to 5mM and EDTA was added to 2mM to chelate Mg2+ and increase the nucleotide exchange rate. Three units of calf intestinal phosphatase was then added to cleave the released GDP molecules, and GDP molecules produced from GTPγs hydrolysis as the sample ages, thereby minimizing the re-formation of yARF1·GDP during NMR studies. This mixture was incubated for over 1 hour and an additional 2mM MgCl2 was added, resulting in a final NMR sample that contains ~0.8mM yARF1, 5mM GTPγs, 10% (w/v) DMPC/DHPC (q=0.25), 3U calf intestinal phosphatase, 10mM K2HPO4-KH2PO4 (pH 7), 50mM NaCl, 10mM K2SO4, 4mM MgCl2, 2mM EDTA, 5% D2O, and 5mM dithiothreitol.
3.2 Data Acquisition and Processing
The 1JNC’, 1JNCα, and 2JNCα couplings were measured for the GTPγs -bound, myristoylated yeast ARF1 protein (2D, 13C, 15N) associated with DMPC/DHPC (q=0.25) bicelles, in isotropic solution and in anisotropic poly-acrylamide gel medium [31]. The myr-yARF1·GTPγs·bicelle complex tumbles at 25°C with an effective rotational correlation time of ~ 30ns [33,31], which corresponds to a molecule weight of ~70 kDa. The experiments were conducted on a 900MHz Varian VNMRS spectrometer equipped with a cold probe. The 15N acquisition time is 35ms. The modulation delays varied from 10ms to 100ms in 10ms steps for all experiments. The number of scans was ramped up as the relaxation delay increased to generate signals of comparable S/N at each point. For isotropic conditions, 8 scans were used for 10 and 20ms modulation delays, 12 for 30 and 40ms, 16 for 50 and 60ms, 20 for 70 and 80ms, 28 for 90 and 100ms. For the anisotropic condition, the number of scans was doubled for the corresponding modulation delays. The isotropic and anisotropic experiments take ~24 and ~48 hours respectively. Data were processed with the software NMRPipe [34]. A cosine square apodization function was applied prior to zero filling and Fourier transformation.
3.2 Error Analysis
The error in IJmod/Iref is propagated from the S/N values of the J-modulated and reference spectra by:
| (3) |
The spectral S/N is defined as the ratio of the peak intensity over the root-mean-square (RMS) of noise. To evaluate the errors of the J couplings, Monte Carlo simulations were conducted where Gaussian noise with a mean of 0 and a deviation of σ (Eq. 3) was added to IJmod/Iref at each time point. The error of J coupling was estimated as the standard deviation of least-squares fitted J couplings from 500 Monte Carlo simulations. The RDC error is the sum of the couplings errors from isotropic and anisotropic media.
The errors of Karplus parameters in Eq. 4 were also estimated by 500 Monte Carlo simulations where Gaussian errors were added to the 1JNCα and 2JNCα couplings.
4. Results
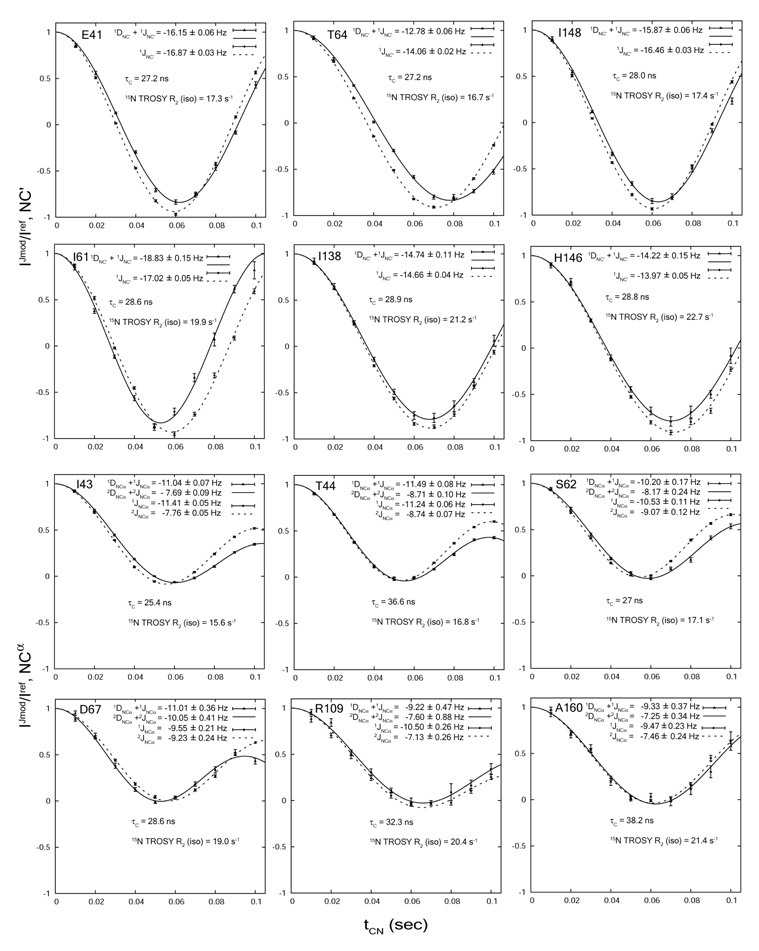
Figure 2 shows representative fits of couplings in isotropic (J) and anisotropic (J+D) media. The rotational correlation time τc at 25° and the 15N TROSY R2 relaxation rates are also indicated for selected residues. For both 1JNC’ and 1JNCα, six representative fits are shown. The upper three fits are selected from those with RDC errors smaller than average, while the lower three fits are from those with RDC errors around average (0.19Hz for 1DNC’, and 0.67Hz for 1DNCα). The trend is clear that the measurement error is positively correlated with 15N TROSY R2. It is also apparent that the RDC error comes mostly from the anisotropic measurement. The τc values indicate that these residues reside in the well-structured part of the protein rather than in the fast re-orientating regions. The J and residual dipolar couplings extracted based on Eq(1) are tabulated in Table 1. Also included in Table 1 are the back-calculated RDC’s based on the crystal structure of the non-myristoylated, GTP-bound mouse ARF1 protein without the N-terminal 17 residues (mARF1-GTP-Δ17) (PDB: 1o3y), which shares 76% sequence identity with the yeast ARF1 protein. The 2DNCα couplings are expected to have a maximum value of only 0.5~0.6 Hz based on the 1DNC’ and 1DNCα couplings, so accurate measurement is inaccessible on this particular system. The measured 1JNC’, 1JNCα and 2JNCα couplings range from 13.2 to 17.3 Hz, 8.3 to 13.1 Hz, and 5.8 to 10.4 Hz, while the typical 15N HSQC and TROSY linewidths in highly digitized spectra are 31 and 25 Hz respectively. Thus, these couplings are not expected to be well resolved using frequency separation experiments.
Figure 2.
Representative fits of intensity ratios for the measurement of 1JNC’ (A) and 1JNCα/2JNCα (B)couplings in isotropic and aligned media. Residual dipolar couplings lead to differential periodicities between the isotropic and aligned conditions.
Table 1.
Scalar and dipolar couplings for yARF1-GTPγS in bicelles.
| 1JNC’ | 1JNCa | 2JNCa | 1DNC’ | 1DNC’ | 1DNCa | 1DNCa | |
|---|---|---|---|---|---|---|---|
| L3 | −14.98±0.01 | −10.07±0.06 | −8.12±0.07 | 0.51±0.02 | * | −0.34±0.13 | * |
| F4 | −14.78±0.02 | −10.38±0.08 | −6.73±0.07 | −0.66±0.04 | * | 0.13±0.17 | * |
| S6 | −14.89±0.02 | −10.54±0.10 | −7.03±0.08 | 0.17±0.06 | * | −1.14±0.21 | * |
| L8 | −14.96±0.02 | −9.88±0.13 | −6.42±0.09 | −0.88±0.04 | * | 0.73±0.27 | * |
| F9 | −14.43±0.02 | −10.32±0.12 | −6.67±0.10 | 0.09±0.05 | * | 0.21±0.26 | * |
| S10 | −14.93±0.02 | −10.73±0.12 | −7.33±0.14 | 0.64±0.08 | * | −0.50±0.35 | * |
| N11 | −15.27±0.02 | −10.72±0.09 | −6.39±0.06 | −0.08±0.03 | * | 0.25±0.19 | * |
| L12 | −15.44±0.02 | −10.02±0.11 | −6.80±0.11 | −0.44±0.04 | * | 0.66±0.22 | * |
| F13 | −14.95±0.02 | −10.70±0.12 | 6.85±0.10 | 0.15±0.05 | * | −0.02±0.24 | * |
| G14 | −15.98±0.02 | −11.89±0.07 | −7.00±0.05 | 0.62±0.04 | * | −0.60±0.14 | * |
| N15 | −15.46±0.01 | −11.93±0.04 | −8.87±0.04 | −0.17±0.02 | * | −0.92±0.06 | * |
| R19 | −14.44±0.03 | −10.40±0.15 | −8.97±0.17 | 1.07±0.10 | 1.05 | −0.43±0.35 | −0.36 |
| I20 | −14.97±0.05 | −11.08±0.23 | −8.29±0.21 | −1.08±0.26 | −0.89 | - | - |
| M22 | −14.30±0.11 | −9.59±0.38 | −9.59±0.39 | - | - | −2.30±0.78 | −1.34 |
| G24 | −15.34±0.21 | - | - | - | - | - | - |
| L25 | −16.22±0.16 | −12.01±0.58 | −9.81±0.60 | - | - | - | - |
| G27 | −14.99±0.04 | −10.70±0.28 | −9.75±0.31 | 0.97±0.17 | 0.68 | 0.70±0.61 | 0.03 |
| A28 | −15.49±0.03 | −10.01±0.19 | −6.35±0.14 | −1.39±0.15 | −0.98 | - | - |
| G29 | −15.19±0.30 | - | - | - | - | - | - |
| K30 | −14.70±0.71 | - | - | - | - | - | - |
| T31 | −14.96±0.09 | −10.85±0.32 | −6.64±0.24 | 0.14±0.41 | 0.24 | - | - |
| V33 | −15.11±0.07 | −10.24±0.75 | −5.76±0.27 | 0.11±0.17 | 0.33 | 0.04±1.29 | −0.55 |
| Y35 | −14.45±0.07 | −10.98±0.33 | −6.59±0.24 | −1.03±0.28 | −0.77 | - | - |
| K36 | −15.47±0.06 | −10.63±0.52 | −6.37±0.35 | −0.05±0.19 | −0.1 | - | - |
| L37 | −14.30±0.05 | −10.62±0.27 | −6.96±0.23 | 0.55±0.24 | 0.27 | - | - |
| L39 | −14.84±0.09 | −9.99±0.42 | −6.63±0.41 | - | - | 0.23±1.67 | 0.51 |
| G40 | −15.34±0.06 | −10.68±0.29 | −6.32±0.20 | 0.30±0.18 | 0.17 | - | - |
| E41 | −16.87±0.03 | −12.58±0.56 | −6.55±0.22 | 0.71±0.08 | 0.69 | −4.27±1.03 | −1.49# |
| I43 | −14.90±0.01 | −11.41±0.05 | −7.77±0.05 | 1.06±0.04 | 0.95 | 0.36±0.12 | 1.1 |
| T44 | −14.59±0.01 | −11.24±0.06 | −8.74±0.07 | 0.41±0.04 | 0.17 | −0.25±0.14 | −0.36 |
| I46 | −15.61±0.02 | −11.97±0.08 | −8.59±0.08 | 0.75±0.09 | 0.91 | 0.35±0.24 | 0.22 |
| T48 | −15.76±0.05 | −10.39±0.31 | −10.38±0.36 | −0.33±0.22 | −0.91 | - | - |
| I49 | −14.76±0.03 | −9.51±0.17 | −7.89±0.22 | 1.19±0.74 | 0.81 | 0.81±0.40 | 0.89 |
| G50 | −14.86±0.24 | −10.44±0.51 | −6.28±0.38 | - | - | - | - |
| F51 | −15.64±0.03 | −12.82±0.07 | −8.89±0.08 | −1.08±0.10 | −0.19 | 0.54±0.23 | 0.72 |
| N52 | −15.34±0.03 | −11.77±0.16 | −9.66±0.20 | 1.22±0.13 | 1.23 | −0.35±0.40 | 0.47 |
| T55 | −15.44±0.02 | −11.55±0.09 | −9.06±0.11 | −0.22±0.07 | −0.92 | −0.06±0.26 | −0.06 |
| V56 | −14.73±0.02 | −10.75±0.12 | −8.43±0.13 | 0.92±0.07 | 0.58 | 1.02±0.31 | 0.83 |
| Q57 | −14.45±0.02 | −11.15±0.09 | −8.61±0.10 | 0.92±0.07 | 0.64 | −0.56±0.19 | −0.23 |
| Y58 | −14.72±0.03 | −10.08±0.27 | −8.96±0.31 | 0.41±0.10 | 0.39 | 0.93±0.49 | 0.72 |
| K59 | −14.84±0.02 | −10.05±0.11 | −8.12±0.13 | 1.33±0.07 | 1.2 | −0.75±0.22 | −0.36 |
| I61 | −17.02±0.05 | −10.88±0.22 | −6.71±0.17 | −1.82±0.20 | −0.92 | 0.73±0.76 | 0.86 |
| S62 | −13.94±0.02 | −10.53±0.11 | −9.05±0.12 | 1.02±0.08 | 0.91 | 0.33±0.28 | 0.29 |
| F63 | −14.56±0.03 | −10.94±0.12 | −8.29±0.14 | −1.76±0.16 | −1.69 | 0.31±0.51 | 0.13 |
| T64 | −14.06±0.02 | −10.35±0.11 | −8.63±0.14 | 1.28±0.08 | 1.13 | 0.81±0.37 | 0.56 |
| D67 | −14.24±0.03 | −9.44±0.21 | −9.36±0.24 | −0.26±0.10 | −0.41 | −1.59±0.57 | −1.77 |
| V68 | −16.47±0.05 | −11.74±0.17 | −7.69±0.14 | 1.07±0.14 | 0.41 | 0.50±0.44 | 0.91 |
| G69 | −13.76±0.08 | - | - | 0.16±0.26 | 0.31 | - | - |
| G70 | −14.26±0.35 | - | - | - | - | - | - |
| Q71 | −15.71±0.12 | −11.67±0.56 | −6.70±0.29 | 0.384±0.35 | 0.45 | - | - |
| D72 | −14.44±0.09 | −10.35±0.37 | −8.97±0.50 | −0.62±0.30 | −0.81 | −0.05±1.24 | 0.61 |
| R73 | −14.47±0.12 | −10.94±0.94 | −7.23±0.61 | 0.79±0.38 | 0.43 | - | - |
| S76 | −14.86±0.10 | −10.40±0.29 | −6.40±0.26 | - | - | −1.52±0.90 | −0.89 |
| R79 | −14.89±0.23 | - | - | - | - | - | - |
| H80 | −14.51±0.12 | −11.31±0.98 | −6.11±0.73 | - | - | - | - |
| Y81 | −15.72±0.17 | −10.63±0.46 | −6.55±0.33 | - | - | - | - |
| Y82 | −15.23±0.08 | −10.40±0.45 | −6.56±0.43 | - | - | 1.41±1.30 | 0.65 |
| N84 | −15.39±0.02 | −11.57±0.10 | −8.19±0.10 | - | - | - | - |
| T85 | −16.12±0.10 | - | - | −2.06±0.28 | −1.7 | - | - |
| E86 | −14.87±0.06 | −10.85±0.22 | −7.97±0.24 | 0.93±0.23 | 1.13 | - | - |
| G87 | −15.85±0.11 | −12.57±0.26 | −7.52±0.25 | −1.45±0.41 | −0.85 | - | - |
| V91 | −14.53±0.11 | −10.64±0.60 | −8.41±0.60 | −0.33±0.26 | −0.01 | −0.82±1.07 | −0.76 |
| D93 | −14.06±0.07 | −9.88±0.26 | −7.71±0.19 | −0.39±0.25 | −0.83 | - | - |
| N95 | −15.46±0.08 | −11.23±0.28 | −6.87±0.25 | 0.30±0.39 | 0.04 | - | - |
| D96 | −15.17±0.05 | −9.49±0.22 | −6.63±0.23 | 0.28±0.22 | 0.15 | 0.12±0.78 | 0.75 |
| R97 | −14.56±0.03 | −10.04±0.15 | −7.93±0.16 | −1.18±0.09 | −0.73 | - | - |
| S98 | −15.29±0.04 | −10.06±0.19 | −6.41±0.14 | 0.19±0.19 | −0.03 | 0.38±0.99 | 0.92 |
| R99 | −15.47±0.04 | −10.05±0.29 | −5.88±0.19 | −1.08±0.18 | −0.91 | −0.99±1.42 | −1.08 |
| I100 | −14.79±0.04 | −10.22±0.24 | −6.14±0.16 | 1.15±0.16 | 1.01 | −0.28±0.95 | −0.73 |
| G101 | −14.82±0.05 | −10.69±0.26 | −6.59±0.19 | −0.07±0.18 | −0.2 | 0.21±1.04 | 0.26 |
| E102 | −15.39±0.04 | −10.79±0.18 | −7.23±0.19 | 0.44±0.11 | 0.31 | 0.38±0.53 | 0.33 |
| R104 | −14.78±0.06 | −10.57±0.18 | −7.22±0.19 | - | - | 0.19±0.67 | −0.1 |
| E105 | −14.80±0.05 | −9.53±0.20 | −6.18±0.15 | −0.15±0.19 | 0.07 | - | - |
| V106 | −15.15±0.04 | −10.30±0.20 | −6.98±0.16 | 0.61±0.11 | 0.49 | −1.35±0.55 | −0.36 |
| Q108 | −14.70±0.05 | −10.57±0.26 | −7.09±0.29 | −0.26±0.17 | −0.29 | - | - |
| R109 | −14.55±0.06 | −10.51±0.26 | −7.13±0.26 | 0.87±0.23 | 0.33 | 1.12±0.74 | 0.64 |
| M110 | −15.12±0.08 | −10.81±0.52 | −7.72±0.62 | 1.94±0.45 | 0.92 | - | - |
| L111 | −14.38±0.19 | - | - | - | - | - | - |
| N112 | −14.59±0.65 | −11.14±0.24 | −6.76±0.19 | −1.80±0.79 | 0.40# | −0.06±0.50 | 0.38 |
| E113 | −16.15±0.03 | −10.91±0.11 | −6.55±0.08 | 0.54±0.08 | 0.34 | −0.22±0.33 | −0.56 |
| D114 | −14.45±0.04 | −9.15±0.16 | −9.14±0.18 | 0.14±0.12 | −0.03 | −2.64±0.37 | −1.44 |
| E115 | −14.22±0.06 | −10.14±0.33 | −6.77±0.31 | 0.44±0.28 | 0.54 | - | - |
| L116 | −15.54±0.07 | −10.93±0.55 | −7.87±0.53 | −0.66±0.30 | −1.02 | 0.03±1.39 | 0.71 |
| R117 | −15.25±0.08 | - | - | - | - | - | - |
| N118 | −15.26±0.11 | −10.70±0.25 | −6.65±0.17 | - | - | 1.13±1.69 | 0.45 |
| A119 | −16.19±0.03 | −11.62±0.15 | −6.59±0.09 | −1.35±0.15 | −0.64 | 1.18±0.67 | 0.02 |
| A120 | −15.16±0.29 | - | - | - | - | - | - |
| W121 | −16.42±0.41 | - | - | - | - | - | - |
| F124 | −13.34±0.17 | −9.54±0.74 | −8.35±0.90 | - | - | - | - |
| A125 | −13.15±0.09 | −8.29±0.38 | −8.26±0.35 | −0.24±0.25 | 0.51 | −2.14±0.84 | −1.73 |
| N126 | −14.87±0.04 | −12.77±0.11 | −8.84±0.12 | 1.00±0.15 | 0.77 | −0.04±0.33 | 0.51 |
| K127 | −15.27±0.04 | −9.98±0.26 | −8.28±0.32 | - | - | - | - |
| Q128 | −14.33±0.09 | −10.16±0.36 | −7.24±0.36 | −0.84±0.36 | 0.06 | - | - |
| D129 | −16.08±0.06 | −10.06±0.30 | −6.59±0.27 | 0.21±0.26 | 0.47 | - | - |
| L130 | −16.09±0.03 | −11.98±0.16 | −6.35±0.09 | 0.88±0.11 | 1.09 | −0.70±1.40 | −1.35 |
| E132 | −15.35±0.03 | −10.40±0.16 | −6.92±0.15 | 1.19±0.11 | 1.26 | 1.20±0.49 | 0.72 |
| A133 | −16.14±0.03 | −11.91±0.85 | −6.11±0.43 | −1.19±0.11 | −1.03 | −2.35±1.29 | −1.61 |
| M134 | −15.51±0.01 | −11.26±0.08 | −9.02±0.09 | 0.53±0.07 | 0.64 | −0.01±0.26 | 0.34 |
| S135 | −15.14±0.02 | −12.79±0.06 | −9.40±0.07 | 0.69±0.12 | 0.88 | - | - |
| A136 | −14.05±0.02 | −10.23±0.10 | −8.39±0.12 | −1.22±0.12 | −1.17 | −0.87±0.34 | −1.23 |
| A137 | −15.19±0.03 | −9.75±0.17 | −6.22±0.14 | - | - | - | - |
| I139 | −14.66±0.04 | −10.26±0.24 | −6.63±0.20 | −0.09±0.16 | −0.2 | 0.80±1.06 | 0.48 |
| E141 | −15.11±0.03 | −10.43±0.22 | −6.31±0.14 | −1.27±0.10 | −1.18 | 0.36±0.68 | 0.28 |
| K142 | −14.86±0.03 | −10.02±0.21 | −6.25±0.16 | 0.02±0.13 | 0.04 | - | - |
| L143 | −15.19±0.06 | −10.40±0.22 | −6.39±0.15 | −1.11±0.24 | −1.34 | −1.05±1.03 | −0.67 |
| G144 | −15.22±0.03 | −10.96±0.16 | −6.92±0.14 | 0.13±0.13 | 0.64 | −0.16±0.48 | −0.04 |
| L145 | - | −9.54±0.16 | −6.15±0.11 | - | - | - | - |
| H146 | −13.97±0.05 | −10.75±0.22 | −6.89±0.21 | −0.26±0.20 | −0.03 | −0.43±0.96 | 0.46 |
| I148 | −16.46±0.03 | −10.52±0.19 | −6.11±0.12 | 0.59±0.09 | 0.21 | −1.74±0.87 | −0.92 |
| R149 | −14.33±0.02 | −11.00±0.09 | −8.19±0.09 | −0.18±0.07 | −0.02 | −0.78±0.22 | −1.2 |
| R151 | −17.30±0.02 | −12.57±0.06 | −6.65±0.03 | 0.12±0.06 | −0.31 | 0.28±0.28 | 0.55 |
| I155 | −15.28±0.02 | −11.35±0.09 | −8.32±0.10 | −0.44±0.08 | −0.59 | −1.15±0.25 | −1.14 |
| A157 | −14.26±0.03 | −10.50±0.16 | −8.54±0.16 | 0.43±0.59 | 0.07 | −1.39±0.34 | −1.32 |
| T158 | - | −12.61±0.11 | −9.17±0.15 | - | - | - | - |
| A160 | −14.74±0.04 | −9.44±0.24 | −7.38±0.24 | - | - | 0.17±0.61 | −0.02 |
| T161 | −14.71±0.10 | −10.49±0.31 | −6.93±0.25 | - | - | −1.01±1.59 | 0.27 |
| S162 | −15.19±0.05 | −10.82±0.31 | −6.37±0.21 | 0.46±0.23 | 0.78 | - | - |
| G163 | −15.96±0.12 | −10.30±0.18 | −5.95±0.12 | 0.22±0.63 | 0.48 | - | - |
| E164 | −16.88±0.05 | −12.17±0.20 | −7.12±0.15 | −0.78±0.29 | −0.86 | 2.21±0.90 | 0.99 |
| G165 | −15.03±0.02 | −9.91±0.11 | −9.90±0.13 | −0.74±0.07 | −0.76 | −0.43±0.39 | 0.16 |
| L166 | −14.87±0.03 | −10.65±0.14 | −7.42±0.15 | 0.60±0.10 | 0.47 | 0.55±0.40 | −0.3 |
| Y167 | −14.13±0.06 | −10.42±0.22 | −6.80±0.19 | 1.27±0.43 | 0.99 | −1.17±1.12 | −1.46 |
| E168 | −15.11±0.05 | −9.88±0.31 | −7.37±0.49 | −0.43±0.24 | −0.75 | 1.06±0.93 | 0.74 |
| G169 | −15.30±0.06 | −11.22±0.25 | −7.07±0.25 | 0.76±0.24 | 0.35 | - | - |
| L170 | −14.87±0.05 | −10.06±0.22 | −7.47±0.26 | 0.00±0.15 | 0.27 | −1.31±0.58 | −1.38 |
| W172 | −15.01±0.04 | −9.79±0.24 | −6.96±0.24 | −1.33±0.32 | −0.77 | - | - |
| S174 | −14.76±0.05 | −10.59±0.26 | −7.50±0.37 | −0.36±0.53 | 0.77# | - | - |
| N175 | −15.39±0.03 | −10.95±0.19 | −6.38±0.12 | 0.02±0.17 | 0.36 | - | - |
| S176 | −15.67±0.04 | −10.76±0.14 | −6.93±0.12 | 0.20±0.20 | −0.18 | 1.48±0.76 | 0.64 |
| L177 | −15.67±0.01 | −10.63±0.09 | −6.07±0.05 | 0.10±0.04 | 0.43 | −0.12±0.26 | 0 |
| K178 | −15.05±0.01 | −10.50±0.05 | −6.81±0.04 | 1.10±0.02 | 1.27 | −0.61±0.10 | −0.95 |
| N179 | −15.02±0.01 | −10.93±0.03 | −7.36±0.03 | −0.46±0.01 | * | −0.01±0.06 | * |
| S180 | −15.70±0.01 | −11.39±0.02 | −7.81±0.02 | 0.32±0.01 | * | 0.42±0.04 | * |
back-calculated RDC values unavailable for the N-terminal residues due to their absence in the crystal structure, and the very C-terminal residues which are flexible and therefore excluded from fitting.
Data unavailable due to poor multiple-point fitting by Eq(2). For the isotropic measurement, fitting quality is deemed “poor” if the Monte Carlo simulation error is greater than 1Hz. RDCs with errors over 2Hz are deemed “poor” and excluded.
RDC data excluded from Figure 3 due to significant deviation from back calculated values.
It is worth noting that the ratio in the 1JNC’ experiment oscillates roughly between 1 and −1, depending on the level of 13C incorporation and inversion efficiency, while the ratio in the 1JNCα-2JNCα experiment oscillates within a much narrower range over the same modulation delays (Figure 2). The smaller dynamic range for the latter experiment makes it more prone to experimental errors. Obviously the equation for the latter experiment also has a significantly longer period than a single cosine function. This is easily seen by rewriting the cos(πJ1t)·cos(πJ2t) function as 1/2·(cos(π(J1+J2)t))+cos(π (J1−J2)t)). Assuming J1 = 11 Hz and J2 = 7 Hz, the slow and fast components have periods of 0.5s and 0.11s respectively. It is impractical to take modulation delays up to the range of the slow modulation without substantial losses in sensitivity. The inability to sample all features of the curve also tends to lower the accuracy. As a result, the measurement accuracy of 1JNC’ is superior to those of 1JNCα and 2JNCα. Errors derived from least-squares fits of 500 Monte Carlo simulations are give in Figure 2 for the residues used as examples. Cases where the simulated errors are greater than 1 Hz for the isotropic measurement are deemed too noisy to produce acceptable fits and are excluded from Table 1. RDC’s with errors greater than 2Hz are also excluded. These filtering steps resulted in a larger number of reliable 1DNC’ couplings than 1DNCα couplings (Table 1). The average measurement errors of 1DNC’ and 1DNCα couplings are 0.19 and 0.67 Hz for the accepted residues. The RMS between the experimental 1DNC’ and 1DNCα couplings and their back-calculated values are 0.36 and 0.55Hz respectively. A small number of accepted residues that deviate from the back-calculated values significantly more than RMS were excluded from Figure 3. These include 2 out of 93 for 1DNC’ (N112 and S174), and 1 out of 71 for 1DNCα (E41) (Table 1). One concern with the measurement of 1JNCα/1DNCα/ and 2JNCα is that for Gly, Ser, and Thr, the 13C inversion efficiency could be degraded due to displacement of their chemical shifts from average values [10]. Here we used a reburp pulse centered on 56ppm with a bandwidth of 50ppm, which covers the entire Cα region (Figure 1, legends). We also explicitly take the inversion efficiency into account during data fitting (Eq. (1)). Based on the experimental and back-calculated 1DNCα values, the effects of Cα chemical shift dispersion do not seem particularly severe (data not shown).
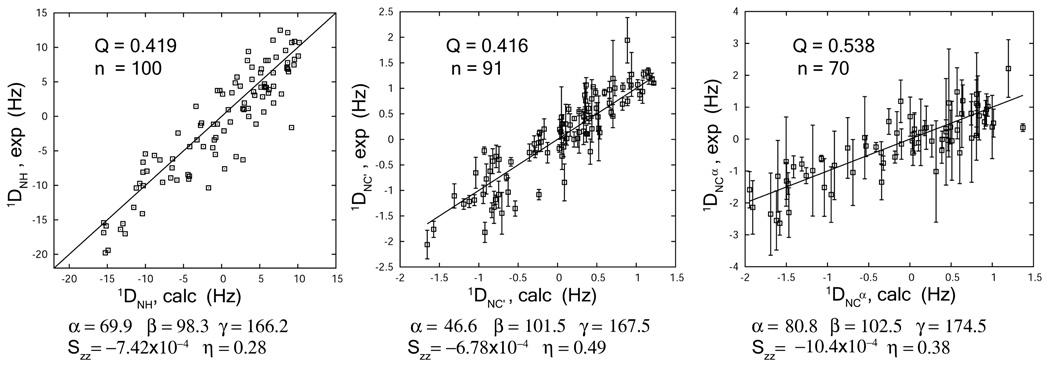
Figure 3.
Best fits of the 1DNC’ and 1DNCα couplings by single value decomposition to the crystal structure of non-myristoylated mARF1-GTP-Δ17. Fit of the backbone amide 1DNH couplings measured from an interleaved pair of TROSY-HSQC experiments and an HNCO-based 3D J-NH experiment is provided as a reference. The alignment tensor parameters are indicated, including the Euler angles (by z-y-z convention) α, β and γ for transforming the given molecular frame into the principle axis frame, the alignment principle order parameter Szz and the asymmetry parameter η (η = (Sxx−Syy)/Szz ).
5. Discussion
Since a high-resolution structure of the bicelle-bound and myristoylated yeast ARF1·GTP is not available, the 1DNC’ and 1DNCα couplings are compared with the back-calculated values based on the crystal structure of the non-myristoylated, GTP-bound mouse ARF1 protein (PDB: 1o3y). These comparisons are presented in Figure 3. In addition to coming from a different organism, the construct used for this crystal structure is missing the N-terminal 17 residues (mARF1-GTP-Δ17) as well as the N-terminal myristoyl chain. However, mouse ARF1 shares 76% sequence identity with the yeast ARF1 protein. RDCs from the N-terminal 17 residues are excluded from Figure 3 due to their absence in the crystal structure. The relatively low Q factors indicate there is significant structural similarity between the two proteins. The deviation from a perfectly linear match is a combined effect of RDC measurement errors, structural differences between yeast and mouse proteins, structural perturbation upon myristoylation, N-terminal deletion, bicelle interaction, and any internal dynamics. Given these potential contributions, Q factors in the 0.4–0.5 range are quite acceptable.
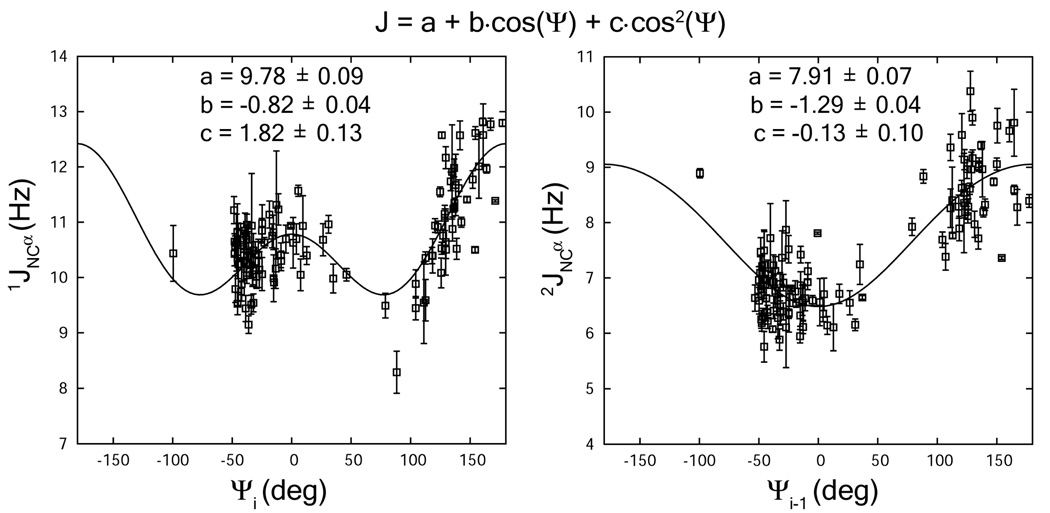
The 1-bond 1JNCα and 2-bond 2JNCα scalar couplings are expected to follow a Karplus equation as a function of the backbone Ψi and Ψi−1 angles. We have used a functional form identical to that preciously presented [10] (Eq 4) and optimized parameters using angles derived from the mARF1-GTP-Δ 17 crystal structure. The fit to the optimized equations are shown in Figure 4. The coefficients as shown in Eq 4 are similar to previously reported values, 9.78±0.09, −0.82±0.04, and 1.82±0.13 for 1-bond 1JNCα, and 7.91±0.07, −1.29±0.04, and −0.13±0.10 for 2-bond 2JNCα [10].
Figure 4.
Fitting of the 1JNCα and 2JNCα couplings to the Karplus relationship as a function of backbone torsion angles Ψi and Ψi−1 respectively. The torsion angles were calculated from the crystal structure of non-myristoylated mARF1-GTP-Δ17.
| (4) |
The agreement and indicated precision of parameters suggests the data will be useful in optimizing backbone Ψi and Ψi−1 torsional angles.
Acknowledgements
This work was supported by grants R01 GM061268 from the National Institute of Genral Medical Sciences and U54 GM074958 from the National Institute of General Medical Sciences in support of the Northeast Structural Gernomics Consortium, a part of the Protein Structure Initiative.
Appendix A
For the 1JNC’ experiment (Fig 1A), the detectable signal at point b resides on the transverse 15N magnetization that is in phase with respect to 13C’:
| (A1) |
Here p is the percentage of 13C incorporation (p = 1 for 100% 13C), e is the inversion efficiency of 13C selective π pulses (e = 1 for 100% inversion), and k is the signal damping due to relaxation. The small relaxation difference between in-phase Nx/y and anti-phase 2Nx/yC’Z is neglected because the longitudinal relaxation of 13C’ is much slower than the transverse relaxation of 15N, and therefore a single damping factor k is applied. Both IJmod and Iref contain 3 terms. The first term (1−p) comes from the 12C’ labeled population whose signal is not modulated by 1JNC’ in either case. The other two terms are associated only with the 13C labeled population. The second term is the only difference between IJmod and Iref, which represents the signal that is modulated in the filtered experiment but not in the reference. The third term containing cos(π·1JNC’·t1) results from incomplete 1JNC’ refocusing during 15N chemical shift evolution from a non-ideal 13C π pulse, which causes p·(1−e)/2 of the total signal to be broadened by 1JNC’ after t1 Fourier transform. When centered on chemical shift, this broadened peak has a normalized height h:
| (A2) |
Here R is the transverse relaxation rate during t1 evolution plus any exponential apodization function added during post-processing.
Through substitution by r, given as follows:
| (A3) |
the expression for the 1JNC’ experiment in Eq.(1) can be derived. It must be mentioned that because h depends on the F1 line-width (Eq. (A2)), the ratio r is different between semi-constant time and constant time experiments. However during our data fitting, the variation of r due to linewidth is neglected so that a single r is used with different tCN values, assuming that (π·J)2 ≪ R2 (h ≈ 1) and that 13C inversion is nearly complete (e ≈ 1). The first assumption is also reasonable because the N-C coupling is quite small and an apodization function is generally used to ramp the data down to zero at the end of a relatively short acquisition time, thereby imposing a large relaxation rate.
For the 1JNCα-2JNCα experiment, the 13C labeling and inversion efficiency for both Cα (ι)and Cα (ι−1) has to be considered. Because the weak J-coupling Hamiltonians commute, a simple recursive relationship is observed with every addition of a J-coupled carbon:
| (A4) |
Here In represents the intensity from a system of n J-coupled carbons. The 13C percentage and the inversion pulse efficiency of the n-th carbon are pn and en. It is simple to solve the recursive relation above to get:
| (A5) |
For n=2, as illustrated by the 1JNCα-2JNCα experiment, the intensity ratio between the filtered and reference experiments adapts the following general form with 5 floating parameters, a, b, c, 1JNCα and 2JNCα:
| (A6) |
Here the coefficients must satisfy a, b, c ≥ 0 and a + b + c ≤ 1. Again, the small effect of line-width variations on the coefficients a, b, and c are neglected. Data fitting by direct application of Eq.(A6) cannot ensure that a, b, and c fall in the legal range. In our actual test, a, b, and c are represented by cosine square functions:
| (A7) |
This works well for most couplings, but the 1DNCα couplings obtained using Eq. (A7), however, agree poorly with the back-calculated values based on the crystal structure of mARF1-GTP-Δ17 (Q factor = 1.24, data not shown). In contrast, the simplified relations, with only 3 floating parameters, shown in the bottom lines of Eq.(1)/(2) significantly improves the agreement (Figure 3). This suggests that in Eq. (A6), the extra degrees of freedom led to over-interpretation of the input data with limited S/N, and trapped the fitting procedure in an erroneous minimum.
In the derivation of the bottom line in Eq. (1), the following restraints are imposed to avoid over-fitting:
| (A8) |
This approximation is quite good when (π·J)2 ≪ R2. Through this approximation, the following equation is derived:
| (A9) |
where,
| (A10) |
Eq. (A9) is further simplified to Eq. (1) (bottom) by requiring r1 ≈ r2 and thus replacing them with a single parameter r. The 13C incorporation level and inversion efficiency usually vary within a small range (i.e., p1 ≈ p2 and e1 ≈ e2), and the small difference in hi associated with the nearly equal 1JNCα and 2JNCα is further scaled down by a small number p·(1-e)/2 (Eq. (A10)). Thus r1 ≈ r2 is a good approximation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Karplus M. Contact electron-spin coupling of nuclear magnetic moments. J. Chem. Phys. 1959;30:11–15. [Google Scholar]
- 2.Bystrov VF. Spin-spin coupling and the conformational states of peptide systems. Prog. Nucl. Magn. Reson. Spectrosc. 1976;10:41–81. [Google Scholar]
- 3.Pardi A, Billeter M, Wüthrich K. Calibration of the angular dependence of the amide proton-Cα proton coupling constants, 3JHNα, in a globular protein: Use of 3JHNα for identification of helical secondary structure. J. Mol. Biol. 1984;180:741–751. doi: 10.1016/0022-2836(84)90035-4. [DOI] [PubMed] [Google Scholar]
- 4.Bax A, Vuister GW, Grzesiek S, Delaglio F, Wang AC, Tschudin R, Zhu G. Measurement of homo- and heteronuclear J couplings from quantitative J correlation. Methods Enzymol. 1994;239:79–105. doi: 10.1016/s0076-6879(94)39004-5. [DOI] [PubMed] [Google Scholar]
- 5.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear magnetic dipole interactions in field-orientated proteins: information for structure determination in solution. Proc. Natl. Acad. Sci. USA. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 7.Prestegard JH, Bougault CM, Kishore AI. Residual dipolar couplings in structure determination of biomolecules. Chem. Rev. 2004;104:3519–3540. doi: 10.1021/cr030419i. [DOI] [PubMed] [Google Scholar]
- 8.Bax A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci. 2003;12:1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delaglio F, Torchia DA, Bax A. Measurement of 15N-13C J couplings in staphylococcal nuclease. 1991;1:439–446. doi: 10.1007/BF02192865. [DOI] [PubMed] [Google Scholar]
- 10.Wirmer J, Schwalbe H. Angular dependence of 1J(Ni,Cαi) and 2J(Ni,Cα(i-1)) couplings constants measured in J-modulated HSQCs. J. Biomol. NMR. 2002;23:47–55. doi: 10.1023/a:1015384805098. [DOI] [PubMed] [Google Scholar]
- 11.Ding K, Gronenborn AM. Protein backbone 1HN-13Cα and 15N-13Cα residual dipolar and J couplings: new constraints for NMR structure determination. J.Am.Chem.Soc. 2004;126:6232–6233. doi: 10.1021/ja049049l. [DOI] [PubMed] [Google Scholar]
- 12.Prestegard JH, Mayer KL, Valafar H, Benison GC. Determination of protein backbone structures from residual dipolar couplings. Methods Enzymol. 2005;394:175–209. doi: 10.1016/S0076-6879(05)94007-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller GA, Choy WY, Yang D, Forman-Kay JD, Venters RA, Kay LE. Global folds of proteins with low densities of NOEs using residual dipolar couplings: application to the 370-residue maltodextrin-binding protein. J. Mol. Biol. 2000;300:197–212. doi: 10.1006/jmbi.2000.3842. [DOI] [PubMed] [Google Scholar]
- 14.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 15.Tolman JR, Prestegard JH. Measurement of amide 15N-1H one-bond couplings in proteins using accordion heteronuclear-shift-correlation experiments. J. Magn. Reson.B. 1996;112:269–274. doi: 10.1006/jmrb.1996.0141. [DOI] [PubMed] [Google Scholar]
- 16.Yang D, Venters RA, Mueller GA, Choy WY, Kay LE. TROSY-based HNCO pulse sequences for the measurement of 1HN-15N, 15N-13CO, 1HN-13CO, 13CO-13Cα and 1HN-13Cα dipolar couplings in 15N, 13C, 2H-labeled proteins. J. Biomol. NMR. 1999;14:333–343. [Google Scholar]
- 17.Puftonen E, Tossavainen H, Permi P. Simultaneous determination of one- and two- bond scalar and residual dipolar couplings between 13C’, 13Cα, and 15N spins in proteins. Magn. Reson. Chem. 2006;44:168–176. doi: 10.1002/mrc.1836. [DOI] [PubMed] [Google Scholar]
- 18.Griesinger C, Søerensen OW, Ernst RR. Two-dimensional correlation of connected NMR transitions. J. Am. Chem. Soc. 1985;107:6394–6396. [Google Scholar]
- 19.Tian F, Losonczi JA, Fisher MW, Prestegard JH. Sign determination of dipolar couplings in field-oriented bicelles by variable angle sample spinning (VASS) J. Biomol. NMR. 1999;15:145–150. doi: 10.1023/a:1008356210291. [DOI] [PubMed] [Google Scholar]
- 20.Billeter M, Neri D, Otting G, Qian YQ, Wüthrich K. Precise vicinal coupling constants 3JHNα in proteins from nonlinear fits of J-modulated [15N-1H]-COSY experiments. J. Biomol. NMR. 1992;2:257–274. doi: 10.1007/BF01875320. [DOI] [PubMed] [Google Scholar]
- 21.Tjandra N, Grzesiek S, Bax A. Magnetic field dependence of nitrogen-proton J splittings in 15N-enriched human ubiquitin resulting from relaxation interference and residual dipolar coupling. J. Am. Chem. Soc. 1996;118:6264–6272. [Google Scholar]
- 22.Vuister GW, Bax A. Quantitative J correlation: a new approach for measuring homonculear three-bond J(HNHα) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc. 1993;115:7772–7777. [Google Scholar]
- 23.Wienk HLJ, Martínez MM, Yalloway GN, Schmidt JM, Pérez C, Rüterjans H, Löhr F. Simultaneous measurement of protein one-bond and two-bond nitrogen-carbon coupling constants using an internally referenced quantitative J-correlated [15N-1H]-TROSY-HNC experiment. doi: 10.1023/a:1022233103990. [DOI] [PubMed] [Google Scholar]
- 24.McFeeters RL, Fowler CA, Gaponenko VV, Byrd RA. Efficient and precise measurement of Hα-Cα, Cα-C’, Cα-Cβ and HN-N residual dipolar couplings from 2D HN-N correlation spectra. J. Biomol. NMR. 2005;31:35–47. doi: 10.1007/s10858-004-6057-y. [DOI] [PubMed] [Google Scholar]
- 25.Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang D, Kay LE. Improved 1HN-detected triple resonance TROSY-based experiments. J. Biomol. NMR. 1999;13:3–9. doi: 10.1023/A:1008329230975. [DOI] [PubMed] [Google Scholar]
- 27.Ottiger M, Bax A. Determination of relative N-HN, N-C’, Cα-C’, and Cα-Hα effective bond lengths in a protein by NMR in a dilute liquid crystalline phase. J. Am. Chem. Soc. 1998;120:12334–12341. [Google Scholar]
- 28.Lerche MH, Meissner A, Poulsen FM, Søerensen OW. Pulse sequences for measurement of one-bond 15N-1H coupling constants in the protein backbone. J. Magn. Reson. 1999;140:259–263. doi: 10.1006/jmre.1999.1820. [DOI] [PubMed] [Google Scholar]
- 29.Nietlispach D. Suppression of anti-TROSY lines in a sensitivity enhanced gradient selection TROSY scheme. J. Biomol. NMR. 2005;31:161–166. doi: 10.1007/s10858-004-8195-7. [DOI] [PubMed] [Google Scholar]
- 30.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell. Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Kahn RA, Prestegard JH. Structure and membrane interaction of myristoylated ARF1. Structure. 2009;17:79–87. doi: 10.1016/j.str.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cierpicki T, Bushweller JH. Charged gels as orientating media for measurement of residual dipolar couplings in soluble and integral membrane proteins. J.Am.Chem.Soc. 2004;126:16259–16266. doi: 10.1021/ja046054g. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Prestegard JH. Direct measurement of dipolo-dipole/CSA cross-correlated relaxation by a constant-time experiment. J. Magn. Reson. 2008;193:23–31. doi: 10.1016/j.jmr.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]