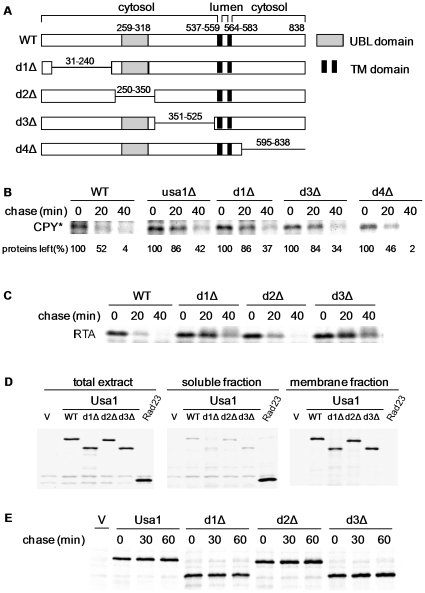
Figure 3. Two N-terminal fragments of Usa1 are important for ERAD.
(A) Domain structure of wild-type Usa1 and various deletion mutants constructed. Two short transmembrane domains (amino acids 537–583) anchor Usa1 to the ER membrane, but most of Usa1 sequences are in the cytosol [10]. Grey box represents the UBL domain and black boxes indicate two transmembrane domains of Usa1. The deleted portions are shown as solid lines. The deleted region in d2Δ is the same as UBLΔ in Figure 2. Deletion strains and plasmids of Usa1 were constructed as described in Experimental procedures. (B) Deletions of two N-terminal segments in Usa1 impair the degradation of CPY*. Pulse chase analysis of CPY* in wild-type and various USA1 mutant strains was performed as described in Figure 2C. The amount of CPY* left (%) is indicated under each lane. (C) Compromised degradation of glycosylated RTA in USA1 deletion mutants. Pulse chase analysis of RTA was done as previously described [17]. (D) Usa1 deletion mutants maintain the ER membrane localization. Yeast cells expressing Flag-tagged Usa1 derivatives were labeled with 35S. Protein extracts were separated into total, membrane and soluble fractions and subsequently immunoprecipitated with Flag-beads. Rad23 was used as a positive control for soluble fraction. (E) Pulse chase analysis of Usa1 wild type and its derivatives. The stabilities of Flag-tagged Usa1 alleles were determined as described in Figure 2C.