Fig. 8.
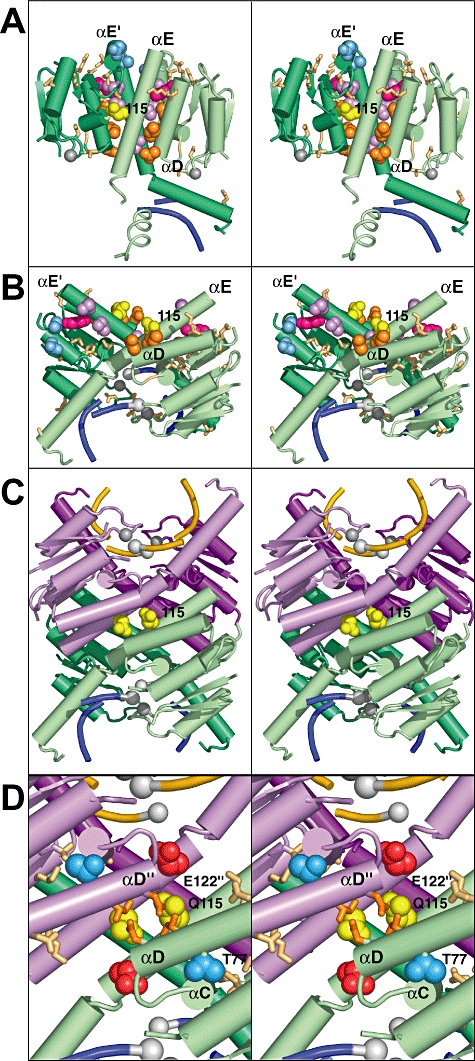
Activating mutations in Sin are located on structural interfaces likely to be involved in assembly of the site I synaptic tetramer. All structures are shown as stereo pairs. A. Crystal structure of the WT Sin dimer bound at site II of resH (pdb: 2R0Q; Mouw et al., 2008). The CTDs (residues 148–202) are omitted. The WT dimer is thought to bind in a similar ‘closed’ conformation at site I, as seen in the crystal structure of WT γδ resolvase bound at site I (pdb: 1GDT; Yang and Steitz, 1995). Side-chains for all residues with activating mutations (Fig. 2A) are highlighted as sticks (light brown) or in spacefill: N72, I75 and V78 (orange; D helix); E98 and V99 (blue; D′ helix), M109, I113 and V120 (violet; E helix); K110 (pink; E helix); Q115 (yellow; E helix). Note that most of the 10 residues shown in spacefill are largely buried in this ‘closed’ dimer, whereas all are exposed on the top surface of the hypothetical ‘open’ dimer shown in (B), i.e. they are at the dimer–dimer interface of the modelled site I synaptic tetramer shown in (C) (the ‘tet’ interface in the last column of Fig. 2A). Grey spheres mark the position of the serine nucleophile (S9) in each subunit. Only 4 bp of the DNA backbone near the centre of site II are shown (blue). B. Hypothetical model of a Sin dimer bound at site I in an ‘open’ conformation, equivalent to one-half of the modelled Sin site I synaptic tetramer shown in (C). Positions of activating mutations are highlighted, as in (A). C. Hypothetical model of a Sin site I synaptic tetramer (the CTDs are not shown). This model is based on the crystal structure of the γδ resolvase site I synaptic tetramer (pdb: 1ZR4): residues 1–119 of each subunit in the γδ resolvase tetramer were replaced by segments of the Sin crystal structure (2R0Q, chain D). Residues 1–102 (core domain) and 103–125 (helix E) of Sin were treated as independent rigid bodies, and fitted to equivalent Cα positions in the γδ resolvase tetramer. The remainder of the γδ resolvase structure (residues 120–183 and the site I DNA) was not altered. Note that the aim was simply to model the likely environment of residues in the Sin synaptic tetramer, including ∼10 Sin residues (96–100 in helix D′ and 103–107 at the N-terminus of helix E) that have no direct equivalent in the γδ structure; no attempt was made to model side-chain conformations. Residue Q115 is highlighted in spacefill (yellow). Only the central 8 bp of site I DNA (blue or yellow backbone, containing a resolvase-induced DSB) are shown; white spheres represent DNA phosphates covalently joined to the serine nucleophile (S10) in 1ZR4. D. Close-up view of Q115 and nearby residues in the modelled Sin site I synaptic tetramer shown in (C). Side-chains are highlighted as in (A) and (B) [except that N72 and I75 are shown as sticks (orange) instead of spacefill]; also shown are E122 (red) and T77 (blue).
