Figure 3. Modes of RNA computation in bacteria based on riboswitches and sRNAs.
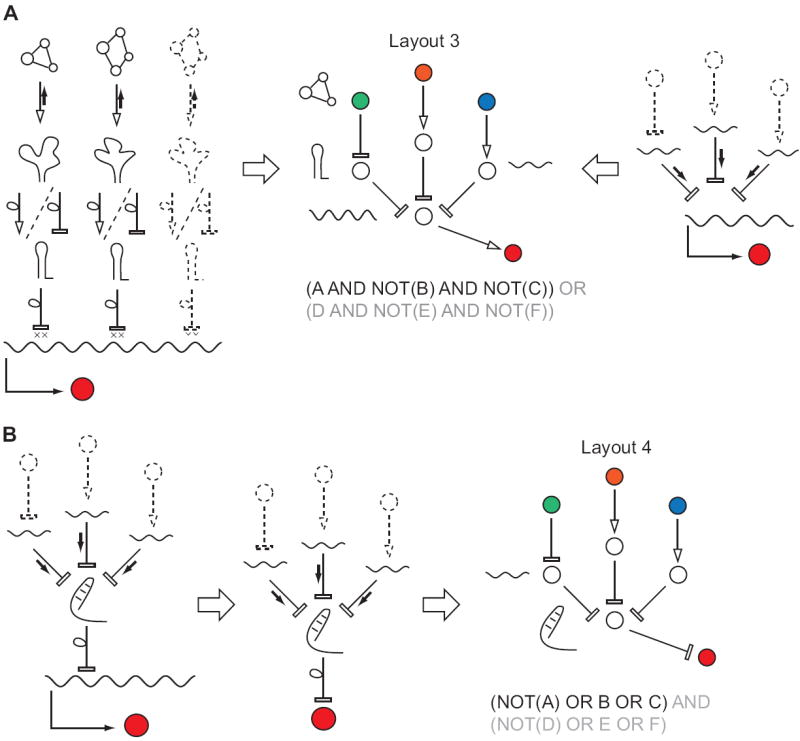
A, A DNF-like computations (central panel) that involve multiple intrinsic terminators controlled by aptamers (left panel) or multiple sRNAs targeting the same transcript (right panel). Depicted diagrams only implement multi-input AND gates, but an OR operation could be included by adding a parallel mRNA expressing the same gene but targeted by a different set of inputs. B, A multi-input OR gate, a portion of a CNF circuit implemented by sRNAs that target a transcription silencing element. Extending this architecture to a full-fledged CNF requires that a number of cis silencing elements control the same mRNA. Such arrangement has not yet been reported but is conceivable.
