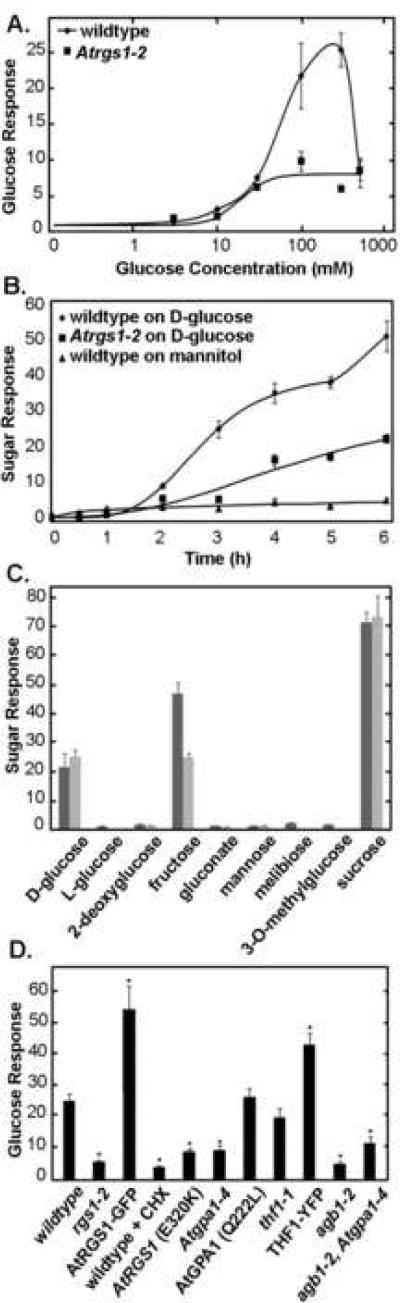
Fig. 1.
Induction of At4g01080 steady-state transcript level (Glucose/Sugar Response). (A) D-glucose dose-dependence of At4g01080 transcript increase by D-glucose. Wild-type and Atrgs1–2 7 day old seedlings were sugar-starved for 2 days and then treated with various concentrations of D-glucose for 3 hours. Wild-type is indicated here as Col-O and Atrgs1 null mutants indicated as rgs1–2. (B) Time dependency of At4g01080 transcript levels in response to treatment with D-glucose or mannitol. Wild-type and Atrgs1–2 7 day old seedlings were sugar-starved for 2 days and then treated with 300 mM D-glucose for varying time periods. (C) At4g01080 transcript level increase in response to a range of sugars and sugar analogues. At4g01080 transcript increase in response to treatment with various sugars and sugar analogues for 3 hours. Dark grey, 100 mM D-glucose; light grey, 300 mM D-glucose. (D) Regulation of At4g01080 transcript level in various genetic backgrounds in response to treatment with D-glucose. Treatment was 300 mM D-glucose for 3 h. rgs1–2: seedlings null for AtRGS1; RGS1-GFP: seedlings null for AtRGS1 and overexpressing an AtRGS1-GFP chimera; WT + CHX: wild-type seedlings treated with cycloheximide (1 or 10 μg/ml) for 1 hour before and during D-glucose treatment; RGS1-E320K: seedlings null for AtRGS1 over-expressing an AtRGS1 mutant containing mutation (E320K) that disrupts the interaction between AtRGS1 and AtGPA1; gpa1–4: seedlings null for AtGPA1; RGS-GFP + gpa1–4: seedlings null for AtRGS1 and AtGPA1 and over-expressing an AtRGS1-GFP chimera; GPA1 (Q222L): seedlings null for AtGPA1 overexpressing an AtGPA1 mutant containing mutation (Q222L), which results in a constitutively active form of the protein; thf1–1: seedlings null for THF1; THF1-YFP: seedlings over-expressing an THF1-YFP chimera; agb1–2: seedlings null for AGB1; agb1–2/gpa1–4: seedlings null for AGB1 and AtGPA1. * indicates values that are significantly different from wild-type plants (p < 0.01). (A–D) Bars or points represent means +/− S.E. After the indicated treatment, RNA was isolated from whole seedlings and used to generate cDNA using oligo dT primers as described in Materials and methods. These cDNA samples were then used for real-time PCR reactions with primers specific for the At4g01080 sequence and the TUB4 sequence (the reference) to determine the level of increase of the At4g01080 transcript level. Each mean is from at least three biological replicates with 3 internal replicates for each to assure precision. For the means presented in panel D, the number of replicates are: Col-O, eleven times1; rgs1–2, eight times; RGS1-GFP, six times; gpa1–4, three times; agb1–2, two times; agb1–2/gpa1–4, three times; GPA1(QL), two times; RGS1(E320K), six times; thf1–1, two times; cyclohexamide, five times.