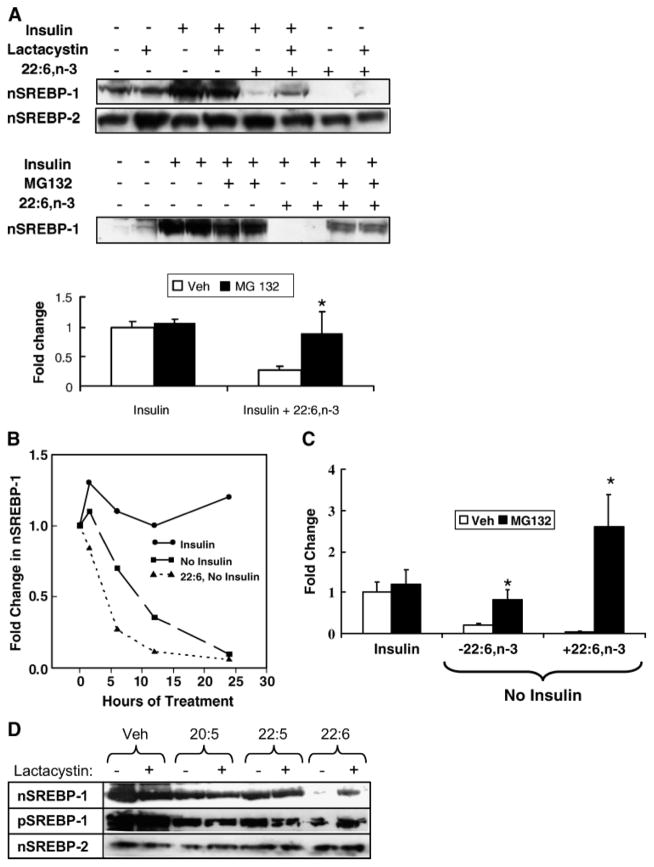
Fig. 4.
Effect of 26S proteasome inhibitors on insulin and 22:6,n-3 regulation of SREBP-1. A: Primary rat hepatocytes were maintained overnight in Williams E medium containing 10 nM DEX and 20 mM lactate but no serum, insulin, or fatty acids. The next morning, cells were switched to medium supplemented with 25 mM glucose in the absence or presence of insulin (10 nM), 22:6,n-3 (100 μM), lactacystin (0.5 μM), or MG132 (10 μM). Lactacystin and MG132 are inhibitors of 26S proteasomal degradation of proteins. In the MG132 experiments, cells were harvested after 12 h of treatment. In the lactacystin studies, cells were harvested after 6 h of treatment. Microsomal and nuclear proteins were assayed for SREBP-1 and SREBP-2 by immunoblotting. Representative immunoblots illustrate the effects of insulin, 22:6,n-3, and lactacystin (upper blot) on nSREBP-1 and nSREBP-2 or MG132 (lower blot) on nSREBP-1. In the absence of insulin or 22:6,n-3, MG132 has no detectable effect on nSREBP-1 abundance. Results of the MG132 experiment are quantified and presented as fold change in nSREBP-1 induced by insulin or insulin + 22:6,n-3 (mean ± SD; n = 4). Veh, vehicle. Results were evaluated by Student’s t-test. * P < 0.001 for vehicle- versus MG132-treated cells. B: Time course of the disappearance of SREBP-1 from hepatocyte nuclei. Primary rat hepatocytes were maintained overnight in Williams E medium containing 25 mM glucose, 10 nM DEX, and 10 nM insulin to induce nSREBP-1. The next morning, the medium was changed to Williams E medium containing 10 nM DEX and 25 mM glucose with insulin, without insulin, or without insulin but with 100 μM 22:6,n-3. Cells were harvested at 1.5, 6, 12, and 24 h for isolation of nuclear proteins. Levels of nSREBP-1 were quantified by immunoblotting. C: Primary rat hepatocytes were maintained overnight in Williams E medium containing 10 nM DEX, 25 mM glucose, and 10 nM insulin to induce nSREBP-1. The next morning, the medium was changed to Williams E medium containing 10 nM DEX and 25 mM glucose with insulin, without insulin, or without insulin but with 100 μM 22:6,n-3. All three groups received vehicle or MG132 at 10 μM. Twelve hours later, cells were harvested for measurement of nSREBP-1 by immunoblotting. Results are expressed as fold change from control (insulin-treated with no MG132) (mean ± SD; n = 3). Results were evaluated by Student’s t-test. * P < 0.01, vehicle- versus MG132-treated cells. D: Cells were treated with insulin in the absence or presence of n-3 PUFAs (100 μM) for 24 h in the absence or presence of lactacystin. Nuclear and microsomal proteins were isolated and assayed for nSREBP-1, pSREBP-1, and nSREBP-2 by immunoblotting. The results are representative of two separate studies.