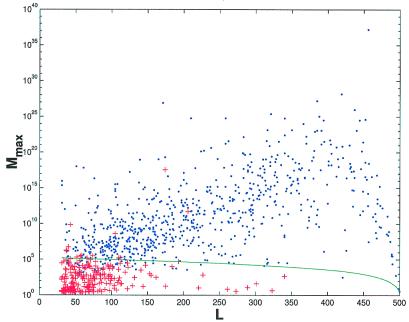
Figure 5.
The maximal number Mmax = 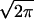 exp((Z/1.2)2/2) of decoys a protein structure calculation can sustain while having the correct native structure recognized (i.e., ɛ > 1.2) vs. the length L of the protein. Blue dots: Mmax vs. L for proteins whose native structure has been recognized in gapless threading. Red crosses: Mmax vs. L for proteins whose native structure has not been recognized. Solid line: the actual number of decoys M obtained by gapless threading as a function of protein length. Notice how well the solid line separates regions of recognized and nonrecognized proteins. This indicates that Mmax is a very good predictor of whether a protein is recognized among a pool of M decoys.
exp((Z/1.2)2/2) of decoys a protein structure calculation can sustain while having the correct native structure recognized (i.e., ɛ > 1.2) vs. the length L of the protein. Blue dots: Mmax vs. L for proteins whose native structure has been recognized in gapless threading. Red crosses: Mmax vs. L for proteins whose native structure has not been recognized. Solid line: the actual number of decoys M obtained by gapless threading as a function of protein length. Notice how well the solid line separates regions of recognized and nonrecognized proteins. This indicates that Mmax is a very good predictor of whether a protein is recognized among a pool of M decoys.