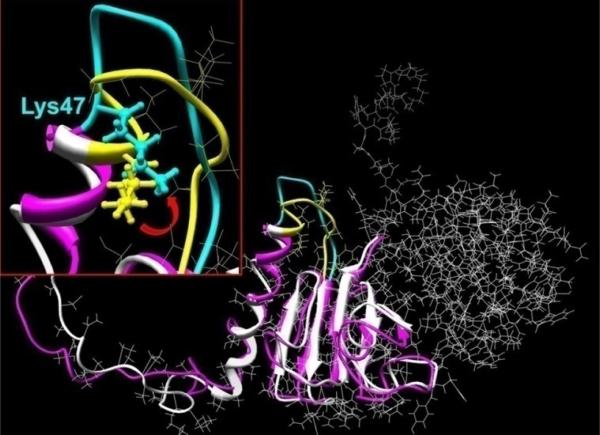
Figure 9. Molecular model the βB1 N-terminal extension.
The murine βB1 crystallin structure was determined using homology modeling and human truncated βB1-crystallin (PDB file: 1oki) as the structural template. Hydrogen atoms were added to the structure which was regularized using OPLS_2005 potentials with 12 Å non-bonded cut-offs. Molecular dynamics in water were performed with the Impact module of the Maestro program software (version 8.0.308: Schrödinger Inc., New York). The initial structure of βB1 monomer is shown in white ribbons and after 20 ps MD equilibration as pink ribbons (details shown for residues 1 to 142). The insert shows structural details of residues 47-57 where the initial and equilibrated structures are indicated by yellow and cyan, respectively. The location of the tryptic cleavage site (between K47V48) is indicated by the ball and stick rendition and the red arrow shows approximate direction of Lys 47 and the peptide movement.