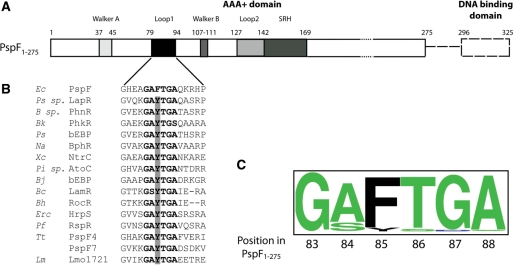
Figure 1.
PspF organization. (A) Full-length PspF consists of the AAA+ domain (residues 1–275, solid box) and a DNA binding domain (residues 296–325, dashed box). Walker A (residues 37–45), L1 loop (residues 79–94), Walker B (residues 107–111), L2 loop (residues 127–142) and the SRH (second region of homology, residues 142–169 with putative arginine fingers R162 and R168) are highlighted in PspF1–275. (B) Protein sequence alignment using the 248 annotated bEBP sequences demonstrated the following microorganisms possess a naturally occurring tyrosine substitution (highlighted by the grey bar) in the ‘GAFTGA’ motif. Ec, Escherichia coli; Ps sp., Pseudomonas sp. KL28; B sp., Burkholderia sp. RP007; Bk, Burkholderia kururiensis; Ps, Pseudomonas syringae pv. Tomato; Na, Novosphingobium aromaticivorans; Xc, Xanthomonas campestris pv. Campestris; Pi sp., Pirellula sp.; Bj, Bradyrhizobium japonicum; Bc, Bacillus cereus ATCC 14579; Bh, Bacillus halodurans; Erc, Erwinia chrysanthemi; Pf, Pseudomonas fluorescens; Tt, Thermoanaerobacter tengcongensis; Lm, Listeria monocytogenes. (C) Conservation of the ‘GAFTGA’ motif residues represented by Weblogo.