Abstract
Human hepatitis delta virus (HDV) is the only animal virus known to replicate its RNA genome using a host polymerase because its only virally encoded proteins, the small and large hepatitis delta antigens (HDAg-S and HDAg-L), lack polymerase activity. Although this makes HDV an ideal model system to study RNA-directed transcription in mammalian cells, little is known about the host factors involved in its replication. To comprehensively identify such host factors, we created a stable cell line carrying a functional FLAG–HDAg-S. Anti-Flag immunopurification and mass spectrometry identified >100 proteins associated with FLAG–HDAg-S, many of which had predicted roles in RNA metabolism. The biological relevance of this screen was strongly supported by the identification of nine out of the 12 subunits of the RNA polymerase II complex thought to mediate HDV replication. To further investigate the significance of these factors for HDV replication, we selected 65 proteins to look for factors that would also affect the accumulation of HDV RNA following siRNA knockdown. Fifteen and three factors were found to regulate HDV RNA accumulation negatively and positively, respectively, upon RNAi knockdown. Our results provide a valuable resource for future research to advance our mechanistic understanding of HDV replication and RNA-directed transcription in mammalian cells.
Keywords: proteomics, RNAi screen, hepatitis delta virus, RNA-directed transcription
INTRODUCTION
Human hepatitis delta virus (HDV) is the smallest known animal virus, with a single-stranded, circular RNA genome of ∼1680 nucleotides in length (for review, see Taylor 2006). It is considered a satellite virus of hepatitis B virus (HBV) because it requires the HBV surface antigen (HBsAg) for virion assembly and transmission. Replication itself, however, does not require HBV proteins. HDV contains only one gene, which encodes for the hepatitis delta antigen (HDAg). The HDAg consists of two distinct isoforms, the small (HDAg-S) and the large (HDAg-L) proteins, both of which lack polymerase activity. HDAg-S is required for replication, while HDAg-L—the result of RNA editing of the HDAg-S stop codon—is dispensable for replication but facilitates viral particle formation. To compensate for its limited protein coding capacity, HDV relies heavily on host factors and its RNA structure. Accumulating evidence supports a model that, once inside the cell, human RNA polymerase II (Pol II) mediates HDV replication by dual rolling-circle transcription from circular genomic and antigenomic RNAs (Macnaughton et al. 2002; Chang et al. 2008), both of which assume compact, unbranched, rod-like structures due to >70% intramolecular base-pair complementarities. As such, HDV is currently the best example of RNA-directed transcription by a nonviral polymerase in vertebrate cells.
Besides the involvement of host RNA Pol II, little is known about other host factors involved in HDV replication. These factors are likely to be critical in ensuring the efficiency and fidelity of HDV replication and may also be critical for our understanding of RNA-directed transcription in mammalian cells in general. Given the central role of HDAg-S in viral replication, we hypothesized that some of these host factors would physically interact with HDAg-S. Previously, a yeast two-hybrid screen was performed to search for interaction partners of HDAg-S, but only one protein, delta antigen interacting protein A (DIPA), was identified (Brazas and Ganem 1996). The limitation of this approach is that yeast does not necessarily reflect the physiological conditions of HDV replication in mammalian cells. Recently, the histone H1e was reported to bind to HDAg-S using the tandem affinity purification (TAP) method followed by mass spectrometry (Lee and Sheu 2008). There are two limitations to this approach. First, the TAP-tagged HDAg-S was not proven to be functional; therefore, host factors only interacting with functional HDAg-S could have been missed in that screen. Second, the traditional, low-throughput method of excising bands for mass spectrometry has limited the discovery to abundant proteins such as histones and ribosomal proteins, and other proteins that are easily detected by staining methods.
An alternative approach to search for HDV host factors employed mass spectrometry analysis following UV cross-linking of HeLa nuclear proteins with an RNA corresponding to the right terminal stem–loop domain of HDV genomic RNA (Greco-Stewart et al. 2006). This identified the polypyrimidine tract-binding protein-associated splicing factor (PSF) as a host factor bound to HDV RNA (Greco-Stewart et al. 2006), although its functional involvement during physiological HDV replication remains unclear. Another proteomic approach was used to analyze global alterations in protein expression in Huh7 cells transiently transfected with HDAg-S, HDAg-L, genomic RNA, and antigenomic RNA, respectively (Mota et al. 2008). A total of 32 proteins were identified using this approach. Whether or not they interact with HDAg and/or affect HDV metabolism was not addressed.
There have been additional reports of host proteins that bind to either HDAg or HDV RNA. PML (Li et al. 2006), nucleolin (Lee et al. 1998), B23 (Huang et al. 2001), and transcription factor YY1 (Huang et al. 2008) have been shown to interact with HDAg and were suggested to affect HDV replication. The Pol I specific factor SL1 (Li et al. 2006) and the negative transcription elongation factor DSIF (Yamaguchi et al. 2001) have also been demonstrated to interact with HDAg, but their effect on HDV replication in vivo is not clear. A double-stranded RNA-activated kinase, PKR, has been shown to bind to HDAg-S (Chen et al. 2002) and HDV RNA (Robertson et al. 1996; Circle et al. 1997), and affect HDV replication (Chen et al. 2002). Finally, GAPDH was reported to bind to HDV RNA (Lin et al. 2000), and ADAR1 was found to edit HDV antigenomic RNA (Wong and Lazinski 2002).
Even with these various efforts, it is likely that many of the essential HDV host factors remain undiscovered. To more comprehensively understand the mechanism of HDV replication, an in vivo, unbiased proteomic interaction screen using HDAg-S as the bait would be invaluable and has been proposed by others (Chang et al. 2005). However, this requires the use of highly specific antibodies suitable for such immunoprecipitation studies. Furthermore, antibodies against the HDAg-S sequence itself may disrupt some of the interactions. Epitope-tagging of HDAg-S may address these concerns but is complicated by the fact that the replicative function of HDAg-S is prone to disruption by such sequence additions (Glenn and White 1991; Chang et al. 1993). In this study, we succeeded in creating a cell line stably expressing a Flag-tagged version of HDAg-S capable of supporting HDV replication. Immunoprecipitation and subsequent mass spectrometry analysis identified more than 100 polypeptides associated with HDAg-S, including nine subunits of Pol II. An RNAi screen further confirmed that a significant portion of these factors have a measurable impact on HDV RNA accumulation.
RESULTS
Identification of HDAg interacting proteins by immunoprecipitation and mass spectrometry
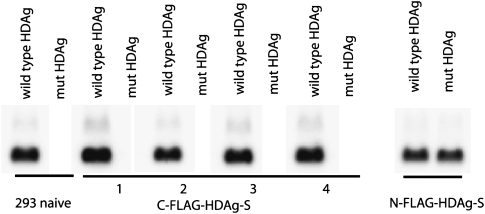
We created a HEK-293-derived cell line stably expressing a Flag-tagged version of HDAg-S, the isoform implicated in viral replication. We chose HEK 293 cells because HDV can replicate efficiently in this cell line, establishing that it has all the necessary host factors for its replication. Moreover, we specifically chose 293 cells as a well-established model system not only for the study of HDV replication per se, but also for possible general RNA-directed RNA transcription. The Flag tag was chosen because it is small and unlikely to interfere with protein–protein interactions and has proven to be very valuable for purifying functional complexes under physiological conditions (Gregory et al. 2006). Both N-terminal and C-terminal Flag tagging were attempted. The N-terminal Flag provided a functional HDAg-S capable of supporting the replication of a replication-deficient mutant virus, which contained an early nonsense mutation in HDAg-S (Fig. 1). C-terminal Flag-tagged cell lines, however, failed to support the replication of this mutant virus (Fig. 1), illustrating the difficulty of retaining activity after slight alterations of this ∼24-kDa protein (Glenn and White 1991).
FIGURE 1.
Functional test of cell lines stably expressing FLAG–HDAg-S (Northern analysis). Cells were transfected with an HDV plasmid with either wild-type HDAg-S, or a mutant version of HDAg-S (mut HDAg) that does not support HDV replication (see Materials and Methods). RNA was isolated 4 d after transfection, and genomic HDV RNA was assessed by Northern blot. Results for four different clones 1–4 made with the same C-terminal Flag-tagged HDAg-S construct and a representative clone made with the N-terminal Flag-tagged HDAg-S are shown.
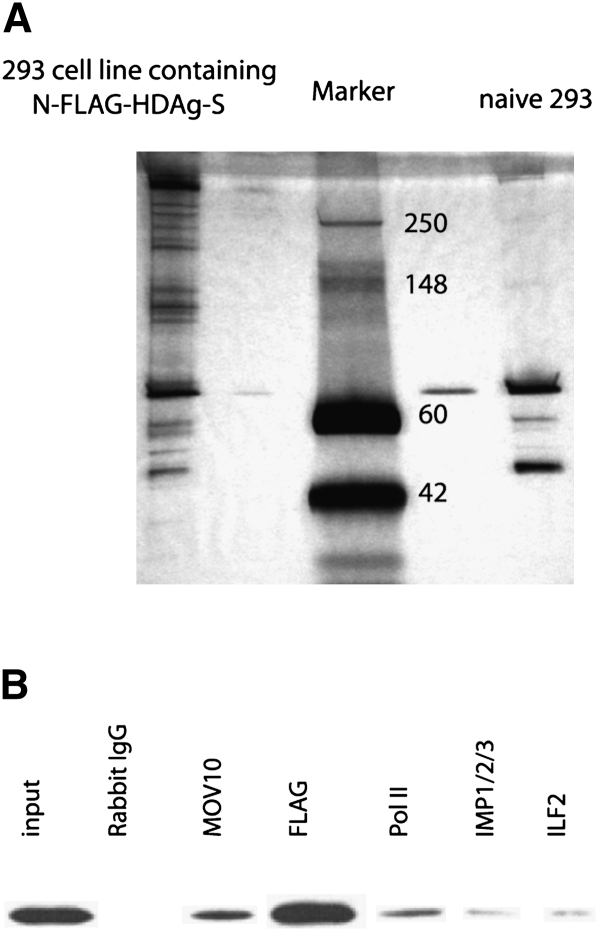
Having established an epitope-tagged HDAg-S functional in viral replication, we isolated associated proteins by anti-Flag immunoprecipitation using naive 293 cells as the negative control. As shown in Figure 2A, the Flag affinity eluates contained a number of polypeptides only associated with HDAg-S, but not present in the control eluate. To eliminate the bias inherent in selecting silver-stained bands for mass spectrometry, to increase sensitivity, and because of the complexity of the staining pattern, the affinity eluate was directly subjected to mass spectrometry analysis by multidimensional protein identification technology (MudPIT) (Delahunty and Yates 2007) without prior selection of individual gel bands. More than 100 proteins associated with HDAg-S were identified, including bait HDAg-S itself and several Pol II subunits (Supplemental Table 1). We note that none of the Pol II subunits had been identified in any previously published screens (see Introduction), although the critical importance of Pol II in HDV replication is well established (Yamaguchi et al. 2001, 2007; Chang et al. 2008). Nine out of the 12 Pol II subunits were identified in this work. Other subunits were not identified, possibly because of their low concentration caused by weak and/or indirect interaction with HDAg-S and/or their small size. Since Pol II has been reported to interact with HDAg-S (Yamaguchi et al. 2001, 2007; Chang et al. 2008), the presence of these Pol II subunits greatly validated our screen and suggested that the affinity eluates were indeed enriched for replication factors. We also note that although Pol I had been implicated in HDV replication based on drug inhibition and HDV localization studies (Modahl et al. 2000; Macnaughton et al. 2002), no Pol I- (or Pol III-) specific subunits were detected. Another validation for this proteomic approach is that SPT5 (part of DSIF) and PKR, two previously identified HDAg-S interacting proteins (Yamaguchi et al. 2001; Chen et al. 2002), were also identified by this approach. Interestingly, hnRNP D and ZNF326, whose abundance was changed as a result of HDAg-S transient transfection in a previous proteomics screen (Mota et al. 2008), were also identified as HDAg-S interacting proteins here.
FIGURE 2.
HDAg-S affinity purification and small-scale reciprocal immunoprecipitation. (A) Purification of N-FLAG–HDAg-S-associated proteins by Flag immunoprecipitation (silver stain of immunoprecipitation eluate). Thirty microliters of the immunoaffinity eluate were resolved on a 4%–20% SDS-PAGE gel and silver-stained (faint bands on both sides of the marker lane are due to overflow from the samples in the neighboring lane). The numbers in the marker lane indicate the size of the marker (in KDa). (B) Small-scale reciprocal immunoprecipitation to confirm the interaction of various proteins with N-FLAG–HDAg-S. Whole rabbit IgG was used as a negative control and anti-Flag antibody as a positive control. Shown is a Flag Western blot of the affinity eluates using the various antibodies for immunoprecipitation as indicated at the top. One percent of the input and 30% of the immunoprecipitation eluates were loaded.
We also identified a number of new proteins with known or predicted roles in RNA metabolism and transcription such as hnRNP proteins, RNA helicases, and several transcription factors that may potentially help recruit Pol II to the HDV RNA template, as well as numerous other RNA-binding proteins and processing factors involved in RNA splicing, export, and degradation (see Supplemental Table 1).
We confirmed some of the FLAG–HDAg-S interaction partners by performing small-scale reciprocal coimmunoprecipitation (Fig. 2B), in which we used various antibodies directed against the HDAg-S interaction partners to test whether they, in turn, would immunoprecipitate HDAg-S. As shown in Figure 2B, FLAG–HDAg-S was indeed coprecipitated by these antibodies.
Several other factors reported to interact with HDAg before were not identified by our mass spectrometry analysis. For PML, SL1, nucleolin, B23, Histone H1e, and YY1, this may be due to the use of different cell lines (HEK293-derived cell line in this study, and Huh7, N1, or COS7 cells in previous studies) and/or the use of different reagents (Lee et al. 1998; Huang et al. 2001; Li et al. 2006; Huang et al. 2008; Lee and Sheu 2008).
RNAi screen for factors affecting HDV replication
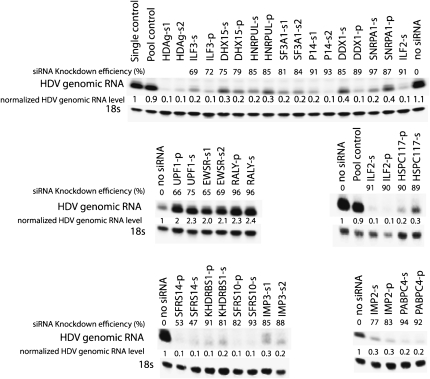
From the above list, we chose 65 genes for further functional study (italicized in Supplemental Table 1) based on their involvement in transcription and RNA metabolism. A pool of siRNAs (Dharmacon) against each gene was transfected into 293 cells to knock down each gene. HDV was introduced the next day by DNA transfection, and RNA was isolated for Northern analysis 48 h thereafter (Fig. 3). The 48-h time point was chosen because the HDV RNA was still accumulating, reaching its highest level in 293 cells by 72 h (data not shown). We examined whether these siRNAs could interfere with HDV genome replication, as assayed by the accumulation of unit-length HDV genomic RNA species. As a positive control, we used siRNAs directed against HDAg-S, which, as reported before (Chang and Taylor 2003), greatly reduced HDV RNA abundance. Most of the pooled siRNAs did not noticeably affect cell growth and viability except, for 10 of them, the cell viability was reduced by >70% as assayed by trypan blue stain. For these targets, we switched to individual “Silencer Select” siRNAs (Ambion) and found that the cell viability was not adversely affected. This suggested that the effect for those 10 pooled siRNAs on cell viability was probably caused by an off-target effect. To reduce the likelihood of false positives due to off-target effects, we monitored the effect on HDV RNA accumulation using individual siRNAs in addition to the siRNA pools (Fig. 3), or using another individual siRNA in the cases where individual siRNAs had already been used in the initial screen (Fig. 3). The effect on HDV RNA accumulation was considered significant only if both pooled siRNAs and at least one individual siRNA, or two different individual siRNAs, had an effect on HDV genomic RNA level by more than twofold. Knockdown efficiencies of the target gene were measured by qRT-PCR (Fig. 3), and 40% was used as a cutoff line for efficient knockdown. Based on these criteria, we found that in addition to the previously reported gene MOV10 (Haussecker et al. 2008), siRNA knockdown for 15 out of the 65 factors significantly decreased the HDV RNA accumulation, while three of them significantly increased the HDV RNA accumulation in two or more separate experiments (Fig. 3).
FIGURE 3.
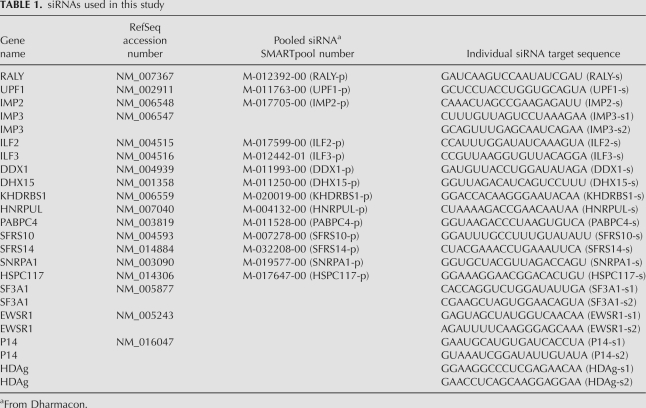
Effect of siRNA knockdown on HDV RNA accumulation in 293 cells (Northern analysis). Numbers represent the relative amounts of 18S-normalized genomic HDV RNA following each knockdown compared to treatment with “Single control,” using a single non-targeting siRNA from Ambion (Catalog number 4390843); or no siRNA control; or “Pool control,” using a pool of non-targeting siRNAs from Dharmacon (set at 1, Catalog number D-001206-13-05). A “p” after each gene symbol indicates that pooled siRNA was used. An “s” after each gene symbol indicates that individual siRNA was used. “s1” and “s2” indicate different individual siRNAs. Shown here are the unit length monomer genomic HDV RNA. Knockdown efficiencies for each target gene were compared to no siRNA control (set at 0%). The sequence or the catalog number of the siRNAs used in this figure is listed in Table 1.
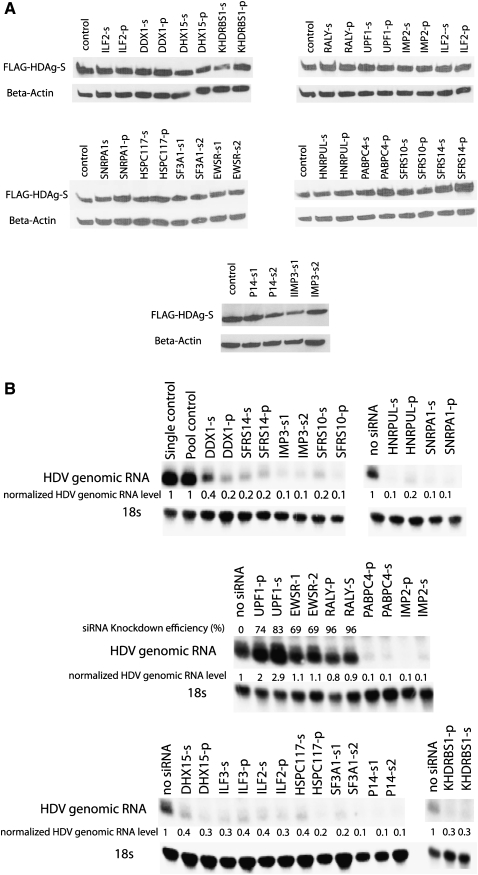
To identify factors that affected HDV RNA levels through their specific role in HDAg mRNA transcription or translation, we performed similar RNAi knockdown experiments in the cells stably expressing N-FLAG–HDAg-S. Importantly, none of these siRNAs showed any significant effect on HDAg-S protein levels, indicating that they did not affect HDAg-S transgene expression derived from the cDNA (Fig. 4A). Notably in these cells, all factors except EWSR1 and RALY still had inhibitory or stimulatory effects on HDV RNA levels similar to those observed in the unmodified 293 cells (Fig. 4B). These data strongly suggest that these factors affect HDV RNA-directed transcription or degradation. Interestingly, EWSR1 and RALY knockdowns did not lead to an increase in HDV genomic RNA levels in this cell line, even though their knockdown efficiencies on the target gene were very similar (Fig. 4B), suggesting that they negatively regulate HDAg-S mRNA expression, possibly via the endogenous HDAg-S mRNA 5′ or 3′ UTRs that were not present in the HDAg-S cDNA construct.
FIGURE 4.
Effect of siRNA knockdown on HDAg-S translation and HDV RNA accumulation in cells stably expressing N-FLAG–HDAg-S. (A) FLAG–HDAg-S level was not noticeably affected by siRNA knockdown as determined by Western blot using anti-Flag antibody. “Control” sample was not transfected with any siRNA. (B) Similar inhibitory or stimulatory effects were observed in cells stably expressing HDAg-S as in naive 293 cells, except for EWSR1 and RALY. Numbers represent the relative amounts of 18S-normalized genomic HDV RNA following each knockdown compared to treatment with non-targeting siRNAs control (=1) or no siRNA control.
DISCUSSION
In this study, we have identified more than 100 proteins associated with HDAg-S, the replicative isoform of HDAg, by mass spectrometry. Among these, we detected nine out of the 12 subunits of Pol II, supporting the premise that the affinity eluates were enriched for factors participating in HDV replication. Importantly, more than 25% of the factors we chose for functional analysis by RNAi knockdown studies had a significant effect on HDV RNA accumulation. Future studies will determine whether these factors directly or indirectly interact with HDAg-S and how they participate in HDV replication. Nevertheless, the associations of the identified candidates with HDAg-S combined with their effect on HDV RNA levels suggest that these are bona fide viral host factors and may be part of the replication machinery.
It is likely that a number of these factors interact with HDAg-S via host RNAs, since they are RNA-binding proteins and our anti-Flag immunoprecipitation was performed without removing RNA:protein interactions. Some of these cellular RNA-mediated interactions may not be important for HDV replication, while some may be relevant for understanding the pathology of HDV infection. One possible way of how host RNA mediate HDAg-S–host protein interactions is through Pol II. Pol II is well known to be associated with a range of RNA processing factors via nascently transcribed RNA and/or the CTD (such as some splicing and transcription related proteins that were also identified in our screen). It is to be expected that at the time when HDAg-S hijacks Pol II, some of these factors will become linked to HDAg-S in an RNase-sensitive manner via Pol II. It is also likely that some of the same factors would also play a vital role in HDV replication. This is supported by the fact that many proteins interacting with FLAG–HDAg-S in our study were also found to interact with Pol II (Das et al. 2007). Some of these proteins had a significant effect on HDV RNA accumulation upon RNAi knockdown, such as ILF2, ILF3, and KHDRBS1, although the RNase sensitivity of their interaction with HDAg-S is yet to be studied.
We have previously reported that RNAi knockdown of one of the HDAg-interacting proteins identified by our approach, MOV10, significantly diminished HDV replication (Haussecker et al. 2008). MOV10 is a putative RNA helicase that had been reported to interact with the Argonaute complex and, consequently, had been suggested to be involved in miRNA-guided gene suppression (Meister et al. 2005). While a function of MOV10 in RNA silencing in humans remains to be identified, it is notable that the Arabidopsis homolog of MOV10, SDE3, is similarly involved in RNA amplification (Dalmay et al. 2001) and that HDV replication is associated with capped small RNAs (Haussecker et al. 2008). This illustrates how factors identified by this screen may be used for further mechanistic study.
Both ILF2 (also known as NFAT45) and ILF3 (also known as NFAT90) were identified in our screen. These two proteins are in the same NF complex and were found to be involved in the replication of another RNA virus, HCV (Isken et al. 2007). ILF2 and ILF3 were shown to bind to the replication signal at the HCV genomic 5′ and 3′ termini and to promote the formation of a loop-like structure of the viral RNA (Isken et al. 2007). It would be interesting to investigate whether they have a similar function in HDV replication by recruiting the HDAg-associated polymerase complex to the HDV hairpin loops that are thought to harbor promoter activity (Beard et al. 1996; Greco-Stewart et al. 2006).
It is not clear these factors affect HDV RNA accumulation directly or indirectly. One interesting thing we noticed is that many factors identified in this screen were also found to be associated with the Argonaute proteins that are involved in miRNA-mediated silencing (Landthaler et al. 2008). This includes the helicase MOV10; the polyadenylae-binding protein 4 (PABPC4); and RNA-binding proteins IMP2, IMP3, RALY, RBM14, ILF2, and ILF3 (Landthaler et al. 2008). AGO4 was also previously identified as a factor affecting HDV RNA accumulation (Haussecker et al. 2008). This suggested a possible link between microRNA function and HDV replication.
We have further identified the two RNA helicases DDX1 and DHX15, which may promote HDV replication by assisting the unwinding of the structurally complex HDV RNA. Several other RNA-binding proteins and processing factors were also identified. The mechanism by which they are involved in HDV replication is not clear and warrants further study.
It remains to be tested whether the factors identified in this screen affect HDV RNA accumulation by affecting transcription or RNA turnover. Our study has laid out a list of candidate proteins for future detailed studies, which should not only give us more insights about how HDV replicates, but possibly more insight into nonviral RNA-directed transcription in vertebrate cells in general. Genome-wide transcription studies have indicated an unanticipated diverse array of transcription and transcripts, including so-called mirror-spliced antisense transcripts (MSATs). MSATs are the reverse complement of spliced mRNA and are best explained as having been generated by RNA-directed transcription from their corresponding mRNA (Cheng et al. 2005). Considering that a single viral protein, HDAg-S, which lacks polymerase activity, is able to facilitate viral replication, and considering the structural diversity of the transcriptome, we believe that nonviral RNA-directed transcription in vertebrate cells is a reasonable hypothesis. Because of its robust replication efficiency and apparent simplicity, HDV may serve as an ideal system for the study of such RNA-directed transcription. By identifying host factors involved in HDV replication, we might learn more about nonviral RNA-directed transcription. For example, HDV-related cellular transcripts may be identified by coimmunoprecipitation with the HDAg-S interaction partners.
Finally, while mass spectrometry-based-interaction analysis and genome-wide RNAi screens in isolation are widely applied to discover viral host factors (Brass et al. 2008; Sessions et al. 2009), we provide a method that couples a medium-scale RNAi screen to a proteomic interaction analysis; therefore, combining evidence for physical interaction with functional requirements in viral replication in a focused and economical format. This allows for the rapid identification of novel targets for antiviral therapy, a particularly pressing problem for newly emerging viruses and viruses with limited patient populations, such as HDV, and for which commercial interest in drug development is low. We suggest that RNAi itself would lend itself to the speedy development of such antiviral therapies.
MATERIALS AND METHODS
Cell culture
To induce HDV replication by DNA transfection, HEK293 cells were transfected with plasmid pCMV3DCHDVx1_2ag expressing 1.2 × unit-length antigenomic HDV RNA (kindly provided by Jeffrey Glenn, Stanford University) with Lipofectamine 2000 (Invitrogen). A plasmid containing a replication-deficient mutant, pCMV3DCHDVx1_2ag_mut, was obtained by introducing a stop and frameshift mutation into pCMV3DCHDVx1_2ag soon downstream from the ATG start codon (atgagccggtccgagtcgaggaa→atgagccggtccgagtAgTagga); therefore, it does not produce functional HDAg-S.
The FLAG–HDAg-S cell line was created by stable transfection of C-terminal or N-terminal Flag-tagged CMV-HDAg-S plasmids with a neomycin marker into HEK293 cells. Cells were selected in 800 μg/mL neomycin/G418 (Invitrogen), and positive clones were screened by anti-Flag Western blot. A Flag Western was performed using Peroxidase conjugated Anti-Flag M2 antibody (Sigma, A8592) and a standard Western protocol. The N-FLAG–HDAg-S was created by cloning the HDAg open reading frame (ORF) into pcDNA3 BamHI and XbaI sites using the following primers: aataGGATCCCAagccggtccgagtcgaggaag and aataTCTAGActatggaaatccccggtttcc. The Flag sequence was inserted between the HindIII and KpnI sites.
Anti-Flag immunoprecipitation
Nuclear extract was made and anti-Flag immunoprecipitation was performed using a previously established protocol (Gregory et al. 2006). Anti-Flag Western blotting was performed to identify the elution fraction with the highest amount of FLAG–HDAg-S, which was subsequently used for silver stain and mass spectrometry. Silver stain was performed using the Pierce SilverSNAP kit (Pierce 24600).
Mass spectrometry
Fifty microliters of Flag-affinity eluate were used for each MudPIT. All samples were analyzed using Mascot (Matrix Science; version 2.1.02), Sequest (ThermoFinnigan; version 27, revision 12), and X! Tandem (http://www.thegpm.org; version 2006.04.01.2). The search was performed using the ipi.HUMAN_v3.25 database assuming the digestion enzyme trypsin. Sequest was searched with a fragment ion mass tolerance of 0 Da and a parent ion tolerance of 2.0 Da. Mascot and X! Tandem were searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 2.0 Da. The iodoacetic acid derivative of cysteine was specified in Mascot, Sequest, and X! Tandem as fixed modifications. Oxidation of methionine was specified in Mascot, Sequest, and X! Tandem as variable modifications.
Scaffold (version Scaffold-01_06_19; Proteome Software Inc.) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at >95.0% probability, as specified by the Peptide Prophet algorithm. Protein identifications were accepted if they could be established at >95.0% probability and contained at least 1 identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Only proteins with 99% probability are listed in Supplemental Table 1. Proteins that were also identified in the 293 naive control were removed in Supplemental Table 1.
TABLE 1.
siRNAs used in this study
Small-scale reciprocal immunoprecipitation
For each immunoprecipitation, 1.5 mg of nuclear lysate were used. The total volume for each immunoprecipitation was brought up to 1 mL using IP buffer (20 mM Tris at pH 8.0, 50 mM KCl, 0.2 mM EDTA, 10% glycerol). Lysate was pre-cleared with protein A/G beads (Pierce, 53133) for 1 h at 4°C before adding 5 μg of antibody. The following antibodies were used: MOV10 (Protein Tech Group, 10370-AP), Flag (Sigma, F3165), Pol II (Abcam, ab5131), IMP1/2/3 (Santa Cruz Biotechnology, SC33594), ILF2 (Abcam, ab28772), and Rabbit IgG (Rockland, 011-0102). The cell lysate was incubated with various antibodies overnight at 4°C, followed by 100 μL of protein A/G beads incubation for 4 h. The beads were washed three times with PBS, followed by three washes with 20 mM Tris (pH 7.8). All of the above procedures were performed at 4°C or on ice. A protease inhibitor (Roche Complete EDTA-Free Tablet) was added into all buffers. Proteins bound to the beads were eluted using 120 μL of 2× Laemmli sample buffer (Bio-Rad) for 5 min at 95°C. A Flag Western was performed using Peroxidase conjugated anti-Flag M2 antibody (Sigma, A8592).
siRNA screen
SMARTpool siRNAs (consisting of four different siRNAs per pool) were obtained from Dharmacon using the catalog numbers in Table 1. Sequences for individual siRNAs are listed in Table 1. Naive 293 cells or cells stably expressing HDAg-S were plated in 24-wells so that they reached 40% confluency the next day for siRNA transfection using 12 nM siRNA and 1 μL of RNAiMAX (Invitrogen). HDV replication was induced by transfecting cells 24 h after RNAi knockdown with 0.4 μg of pCMV3DCHDVx1_2ag (for 293 naive cells) or pCMV3DCHDVx1_2ag_mut (in cells stably expressing HDAg-S) using 1 μL of Lipofectamine 2000 (Invitrogen). Total RNA was harvested 48 h after HDV transfection with Trizol (Invitrogen), and HDV RNA abundance was analyzed by Northern blot and phosphorimage quantitation. Cell extracts were prepared using M-PER mammalian protein extraction reagent (Pierce 78501), and Western blotting was performed using Peroxidase conjugated anti-Flag M2 antibody (Sigma, A8592) first, stripped with Western Stripping Buffer (Thermo Scientific 46430) afterward, and then re-blotted using anti-β actin antibody (Sigma, A5316 and A9917).
Northern hybridization
Northern hybridization was performed using a standard Northern protocol. For the detection of full-length genomic HDV RNAs, 3 μg of Trizol total RNA were separated on 1% denaturing formaldehyde-agarose gels, transferred by wet transfer onto Hybond-N nitrocellulose (Amersham), and hybridized to αUTP32-labeled, T7 polymerase transcribed RNA from BamHI-linearized pCMV3DCHDVx1_2ag. 18S was probed using a γATP32-labeled oligo probe (cctattccattattcctagctgcggtatcc).
qRT-PCR
Total RNA was extracted using Trizol. cDNA was generated using Superscript III (Invitrogen) using the primers listed in Supplemental Table 2. Specific cDNAs were quantitated by quantitative PCR using primers in Supplemental Table 2 and QuantiTect SYBR Green PCR kit (QIAGEN) on a Corbett Rotor-Gene 6000 Real Time PCR system. The relative amount of target gene mRNA was normalized to β-actin mRNA. Specificity was verified by melting curve analysis.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank T. Storm for technical assistance, J. Glenn (Stanford) for reagents, and Andrew W. Guzzetta and the Stanford University Mass Spectrometry laboratory for MudPIT analysis. This work was supported by grant DK78424. from the National Institutes of Health (to M.A.K.), a Helen Hay Whitney Foundation Postdoctoral Research Fellowship to D.C., and a Stanford Dean's Postdoctoral Fellowship to D.H.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1782209.
REFERENCES
- Beard MR, MacNaughton TB, Gowans EJ. Identification and characterization of a hepatitis delta virus RNA transcriptional promoter. J Virol. 1996;70:4986–4995. doi: 10.1128/jvi.70.8.4986-4995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Brazas R, Ganem D. A cellular homolog of hepatitis delta antigen: Implications for viral replication and evolution. Science. 1996;274:90–94. doi: 10.1126/science.274.5284.90. [DOI] [PubMed] [Google Scholar]
- Chang J, Taylor JM. Susceptibility of human hepatitis delta virus RNAs to small interfering RNA action. J Virol. 2003;77:9728–9731. doi: 10.1128/JVI.77.17.9728-9731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MF, Sun CY, Chen CJ, Chang SC. Functional motifs of delta antigen essential for RNA binding and replication of hepatitis delta virus. J Virol. 1993;67:2529–2536. doi: 10.1128/jvi.67.5.2529-2536.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Gudima SO, Tarn C, Nie X, Taylor JM. Development of a novel system to study hepatitis delta virus genome replication. J Virol. 2005;79:8182–8188. doi: 10.1128/JVI.79.13.8182-8188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Nie X, Chang HE, Han Z, Taylor J. Transcription of hepatitis delta virus RNA by RNA polymerase II. J Virol. 2008;82:1118–1127. doi: 10.1128/JVI.01758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CW, Tsay YG, Wu HL, Lee CH, Chen DS, Chen PJ. The double-stranded RNA-activated kinase, PKR, can phosphorylate hepatitis D virus small delta antigen at functional serine and threonine residues. J Biol Chem. 2002;277:33058–33067. doi: 10.1074/jbc.M200613200. [DOI] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Circle DA, Neel OD, Robertson HD, Clarke PA, Mathews MB. Surprising specificity of PKR binding to delta agent genomic RNA. RNA. 1997;3:438–448. [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Delahunty CM, Yates JR., III MudPIT: Multidimensional protein identification technology. Biotechniques. 2007;43:563–569. [PubMed] [Google Scholar]
- Glenn JS, White JM. trans-Dominant inhibition of human hepatitis delta virus genome replication. J Virol. 1991;65:2357–2361. doi: 10.1128/jvi.65.5.2357-2361.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco-Stewart VS, Thibault CS, Pelchat M. Binding of the polypyrimidine tract-binding protein-associated splicing factor (PSF) to the hepatitis delta virus RNA. Virology. 2006;356:35–44. doi: 10.1016/j.virol.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Shiekhattar R. MicroRNA biogenesis: Isolation and characterization of the microprocessor complex. Methods Mol Biol. 2006;342:33–47. doi: 10.1385/1-59745-123-1:33. [DOI] [PubMed] [Google Scholar]
- Haussecker D, Cao D, Huang Y, Parameswaran P, Fire AZ, Kay MA. Capped small RNAs and MOV10 in human hepatitis delta virus replication. Nat Struct Mol Biol. 2008;15:714–721. doi: 10.1038/nsmb.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WH, Yung BY, Syu WJ, Lee YH. The nucleolar phosphoprotein B23 interacts with hepatitis delta antigens and modulates the hepatitis delta virus RNA replication. J Biol Chem. 2001;276:25166–25175. doi: 10.1074/jbc.M010087200. [DOI] [PubMed] [Google Scholar]
- Huang WH, Mai RT, Lee YH. The transcription factor YY1 and its associated acetyltransferases CBP and p300 interact with hepatitis delta antigens and modulate the hepatitis delta virus RNA replication. J Virol. 2008;82:7313–7324. doi: 10.1128/JVI.02581-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Baroth M, Grassmann CW, Weinlich S, Ostareck DH, Ostareck-Lederer A, Behrens SE. Nuclear factors are involved in hepatitis C virus RNA replication. RNA. 2007;13:1675–1692. doi: 10.1261/rna.594207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CZ, Sheu JC. Histone H1e interacts with small hepatitis delta antigen and affects hepatitis delta virus replication. Virology. 2008;375:197–204. doi: 10.1016/j.virol.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Lee CH, Chang SC, Chen CJ, Chang MF. The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting. J Biol Chem. 1998;273:7650–7656. doi: 10.1074/jbc.273.13.7650. [DOI] [PubMed] [Google Scholar]
- Li YJ, Macnaughton T, Gao L, Lai MM. RNA-templated replication of hepatitis delta virus: Genomic and antigenomic RNAs associate with different nuclear bodies. J Virol. 2006;80:6478–6486. doi: 10.1128/JVI.02650-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SS, Chang SC, Wang YH, Sun CY, Chang MF. Specific interaction between the hepatitis delta virus RNA and glyceraldehyde 3-phosphate dehydrogenase: An enhancement on ribozyme catalysis. Virology. 2000;271:46–57. doi: 10.1006/viro.2000.0302. [DOI] [PubMed] [Google Scholar]
- Macnaughton TB, Shi ST, Modahl LE, Lai MM. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J Virol. 2002;76:3920–3927. doi: 10.1128/JVI.76.8.3920-3927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel Argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Modahl LE, Macnaughton TB, Zhu N, Johnson DL, Lai MM. RNA-dependent replication and transcription of hepatitis delta virus RNA involve distinct cellular RNA polymerases. Mol Cell Biol. 2000;20:6030–6039. doi: 10.1128/mcb.20.16.6030-6039.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota S, Mendes M, Penque D, Coelho AV, Cunha C. Changes in the proteome of Huh7 cells induced by transient expression of hepatitis D virus RNA and antigens. J Proteomics. 2008;71:71–79. doi: 10.1016/j.jprot.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Robertson HD, Manche L, Mathews MB. Paradoxical interactions between human delta hepatitis agent RNA and the cellular protein kinase PKR. J Virol. 1996;70:5611–5617. doi: 10.1128/jvi.70.8.5611-5617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JM. Structure and replication of hepatitis delta virus RNA. Curr Top Microbiol Immunol. 2006;307:1–23. doi: 10.1007/3-540-29802-9_1. [DOI] [PubMed] [Google Scholar]
- Wong SK, Lazinski DW. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc Natl Acad Sci. 2002;99:15118–15123. doi: 10.1073/pnas.232416799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Filipovska J, Yano K, Furuya A, Inukai N, Narita T, Wada T, Sugimoto S, Konarska MM, Handa H. Stimulation of RNA polymerase II elongation by hepatitis delta antigen. Science. 2001;293:124–127. doi: 10.1126/science.1057925. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Mura T, Chanarat S, Okamoto S, Handa H. Hepatitis delta antigen binds to the clamp of RNA polymerase II and affects transcriptional fidelity. Genes Cells. 2007;12:863–875. doi: 10.1111/j.1365-2443.2007.01094.x. [DOI] [PubMed] [Google Scholar]