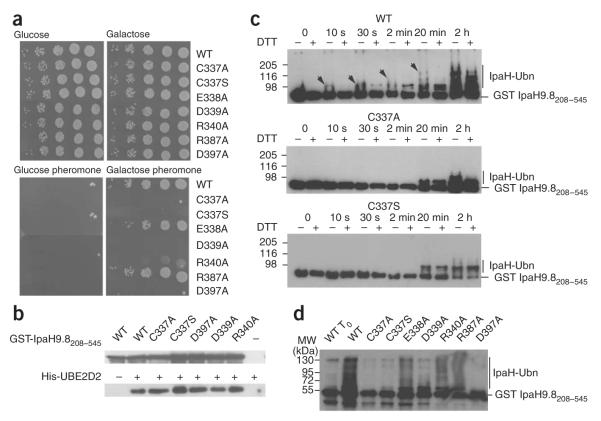
Figure 6.
Functional analysis of conserved residues. (a) Growth on glucose- or galactose-containing plates of serial dilutions of sst2Δ yeast cells harboring plasmids encoding indicated proteins (WT, wild-type IpaH9.8-Flag; C337A; IpaH9.8 in which Cys337 is replaced by alanine (the same nomenclature is used for the other variants)). Below, plates containing α-factor are shown. (b) Immunoblot analysis with anti-IpaH (above) and anti-His (below) antibodies of eluates from glutathione beads added to GST-IpaH9.8208–545 or its C337A, C337S, D397A, D339A and R340A variants incubated with His6-UBE2D2. (c) Immunoblot analysis using anti-IpaH antibodies of reactions performed in the presence of ATP, ubiquitin, E1, UBE2D2, GST-IpaH9.8208–545 (WT, above) or its C337A (middle) and C337S (below) variants. Samples were incubated at 25 °C for the indicated time and treated (+) or not treated (-) with DTT before loading. Arrows indicate the DTT-sensitive species. (d) Immunoblot analysis using anti-IpaH antibodies of reactions performed in the presence of ATP, ubiquitin, E1, UBE2D2, GST-IpaH9.8208–545 or the GST-IpaH9.8208–545 variants carrying the indicated replacement. Samples were incubated at 25 °C for 30 min (except for WT To, which represents the degree of ubiquitination of the wild-type protein at t = 0 min) and treated with DTT before loading.