Fig. 2.
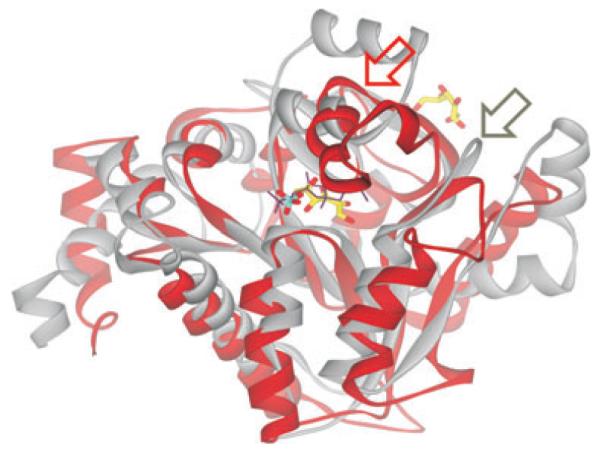
Superimposition of C-CggR and GlcN6P deaminase NagB structures Ribbon representation of C-CggR (gray) and NagB (red) structures. FBP bound to C-CggR is shown as black lines, and the reaction product fructose-6-phosphate (F6P) bound to NagB (PDB code 1FQO, Rudino-Pinera et al., 2002) is shown as a stick model with carbons in yellow, oxygen atoms in red and phosphorous in cyan. Note that the NagB structure contains a second F6P molecule that is bound to its allosteric site. Regions that undergo structural rearrangements upon substrate and ligand binding are indicated by red and gray arrows for NagB and C-CggR respectively.
