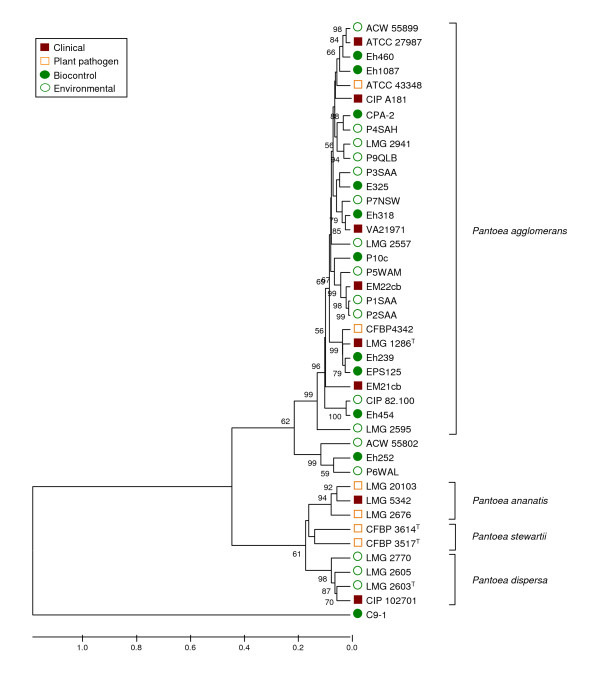
Figure 4.
Clustering of P. agglomerans sensu stricto strains based on UPGMA analysis of concatenated fAFLP patterns obtained using four selective primer combinations. For each strain, a total of 885 data points (bands) indicating the presence (1) or absence (0) of an fAFLP peak were used in the analysis. P. ananatis, P. stewartii and P. dispersa strains were used as reference. Bootstrap values after 1000 replications are expressed as percentages. T: type strain; LMG: Culture Collection, Laboratorium voor Microbiologie, Ghent, Belgium; CFBP: Collection Française de Bactéries Phytopathogènes INRA, Angers, France; CIP: Collection de l'Institut Pasteur, Paris, France; ATCC: American Type Culture Collection, Manassas VA, U.S.A; ACW: Agroscope Changins-Wädenswil, Wädenswil, Switzerland (own strains).