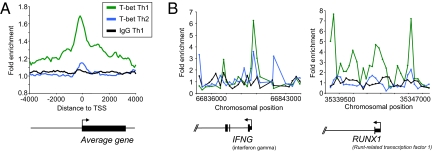
Fig. 1.
T-bet associates with many genes in Th1 cells. (A) Composite T-bet enrichment profile in Th1 cells for genes that show significant binding within 4 kb from their transcription start site (green), compared with the average binding profile for T-bet at the same genes in Th2 cells (blue) and an IgG control antibody ChIP in Th1 cells (black). The plot shows average fold-enrichment (normalized signal from ChIP-enriched DNA divided by the signal from whole cell extract (WCE) DNA. The start and direction of transcription of the average gene is indicated by an arrow. (B) T-bet ChIP signals at IFNG and RUNX1 in Th1 and Th2 cells. The plots show unprocessed enrichment ratios for all probes within a genomic region (ChIP vs. whole genomic DNA) for T-bet in Th1 cells (green), T-bet in Th2 cells (blue), and for an IgG control in Th1 cells (black). Chromosomal positions are from NCBI build 35 of the human genome. Genes are shown to scale below and aligned with the plots by chromosomal position (exons are represented by vertical bars, the start and direction of transcription by an arrow).