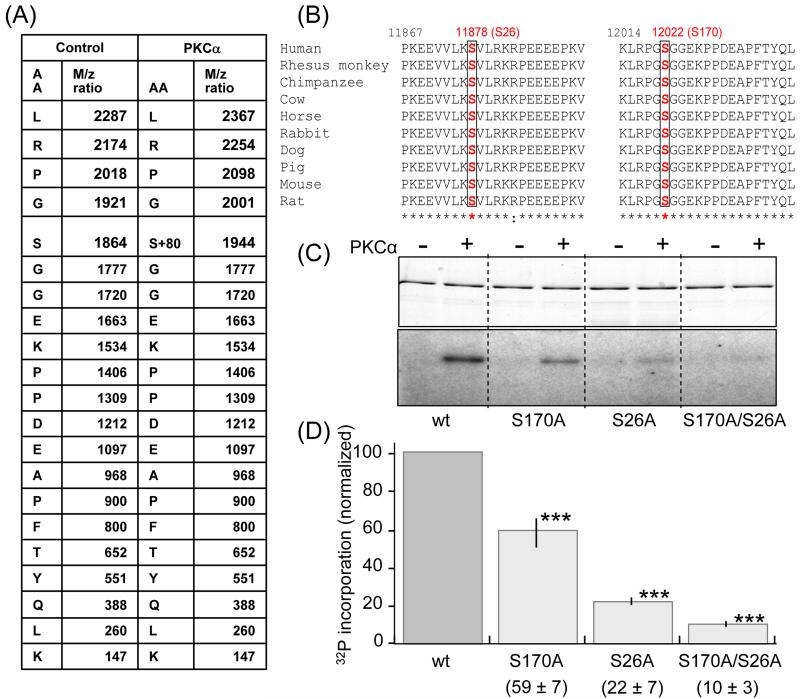
Figure 5. Identification of target PKCα phosphorylation sites on PEVK.
Mass spectrometry was carried out on nonphosphorylated (control) and PKCα phosphorylated PEVK (plus flanking Ig domains). Samples were electrophoresed, followed by in-gel tryptic digestion and analysis by MS/MS. A) Of the 53 identified digestion fragments only fragment LRPGSGGEKPPDEAPFTYQLK had a differential mass of 80 × n Dalton; its fragmentation table with its amino acid sequence and the m/z ratio are shown. The control fragment has a molecular mass of 1863.9 dalton and the phosphorylated fragment 1943.9 dalton (1863.9+80). The fragmentation table shows that the phosphorylated residue corresponds to serine 170 in the PEVK sequence (human N2B cardiac isoform) or S12022 in the full human sequence (Swiss Prot:Q8wz42). (B) Multiple sequence alignments from various species show that Serine170 (red) and its flanking sequence are conserved in all species. Serine 26 (S11878 in the full human sequence) is a conserved serine that was not included in the digestion fragments. (C) In vitro phosphorylation by PKCα kinase of wildtype PEVK, the S170A, S26A and S170/S26A mutants. (Top: Coomassie blue stain gel. Bottom: Autoradiograph). A reduction in 32P incorporation in the mutants can be visibly detected and quantification (D) shows that relative to the wildtype the reduction is highly significant. ***: Significant vs. WT (P<0.001). (n = 9 for wt, S170A, and S170/S26A mutants; n= 5 for S26A).