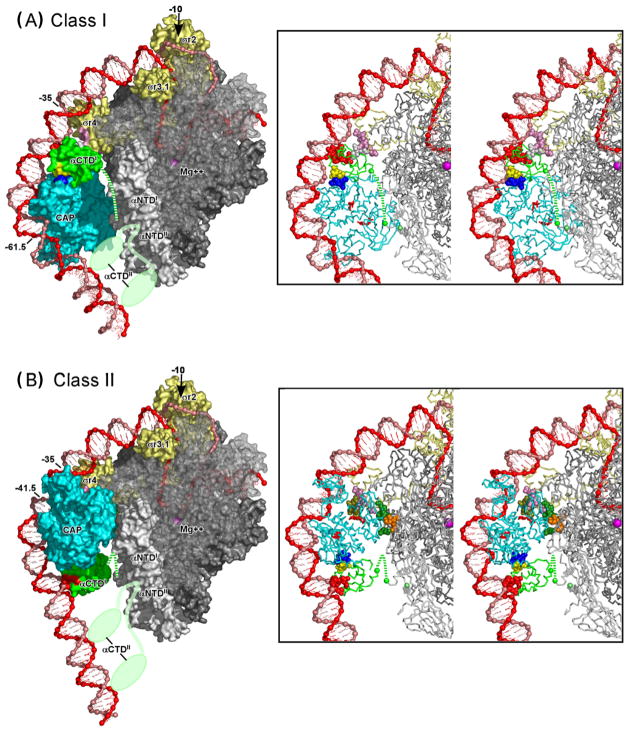
Figure 5.
Transcription activation by CAP: structural models of intact Class I and Class II CAP-RNAP-promoter complexes.
(A) Structural model of the intact Class I CAP-RNAP-promoter complex at lac.
(B) Structural model of the intact Class I CAP-RNAP-promoter complex at CC(−41.5).
In each panel, a molecular surface representation is shown at left; and a stereodiagram with a ribbon representation is shown at right. Colors of CAP and RNAP are as in Fig 3: CAP is in cyan; αCTDI is in green; αCTDII is in light green (shown in two alternative positions in surface representations; omitted for clarity in ribbon representations); σ70 is in light yellow; αNTDI and αNTDII are in light gray; β is in medium gray (semi-transparent in surface representations, to permit view of DNA strands in RNAP active-center cleft); and β’ and ω are in dark gray. Colors of determinants of CAP and RNAP also are as in Fig 3: AR1, AR2, and AR3 of CAP are in dark blue, dark green, and olive green; the 287, 265, and 261 determinants of αCTDI are in yellow, red, and white; the 162–165 determinant of αNTDI is in orange; and the 593–604 determinant of σ70 is in pink. The DNA template and nontemplate strands are in red and pink. The C-terminus of αNTDI (green) the C-terminus of αNTDII (light green), and the active-center Mg++ (magenta) are indicated by spheres. The linker connecting αCTDI and αNTDI is indicated by a dashed green line. The linker connecting αCTDII and αNTDII is indicated in each of two alternative positions as a light green line.
Methods: Models were constructed by: (i) joining crystal structures of the CAP-αCTD-DNA complex (PDB 1LB2 [44**]), the σR4-(−35 element) complex (PDB 1KU7 [47**]), and an RNAP-DNA complex (PFB 1L9Z [49**]; residues 150–160 and 164–170 of αNTDI modelled as in PDB 1BDF [65]; residues 161–163 of αNTDII modelled along shortest sterically allowed path; side chains modelled using MaxSprout [http://www.ebi.ac.uk/maxsprout/] )--superimposing DNA segments of the three structures onto a single, continuous DNA segment having sites spaced as at lac (panel A) or CC(−41.5) (panel B); (ii) deforming conformations of DNA positions −13 to −31 and −41 to −36 (panel A) or −13 to −30 and −38 to 33 (panel B) to minimize the elastic energy of DNA at the base-pair level [50] while satisfying DNA anchoring conditions, non-interpenetration constraints (Cα-Cα distance ≥3.5 Å for all residue pairs), and proximity constraints (Cα-Cα distance ≤12 Å for residue pairs specified below); and (iii) modelling DNA template-strand positions −11 to +20 and nontemplate-strand positions −7 to +20 as in published models of the RNAP-promoter open complex [39,51**]. For panel A, the following proximity constraints were used: proximity of residues 257, 258, 259, and 261of αCTD to at least one of residues 593, 596, 597, 600, 601, and 604 of σR4, and vice versa (mutational analysis [36,37,40**]); and proximity of residue 261of αCTD to residues 596 and 600 of sR4 (suppression analysis [40**]) (residues numbered as in E. coli RNAP). For panel B, the following proximity constraints were used: proximity of residues 19, 21, 96, and 101 of the downstream CAP subunit to at least one of residues 162, 163, 164, and 165 of αNTDI, and vice versa (mutational analysis [56]); proximity of residues 52, 53, 54, 55, and 58 of the downstream CAP subunit to at least one of residues 593, 596, 597, 599, and 603 of σR4 and vice versa (mutational analysis [58,60]); and proximity of residue 58 of the downstream CAP subunit to residue 596 of σR4 (suppression analysis [59]) (residues numbered as in E. coli RNAP). The models have been deposited in the PDB (PDB **** and ****). Figures were prepared using PyMol [http://www.pymol.org]. The view orientation reflects rotation by −45 on the y-axis relative to the “upstream” view orientation in published models of the RNAP-promoter open complex [39,51**]).