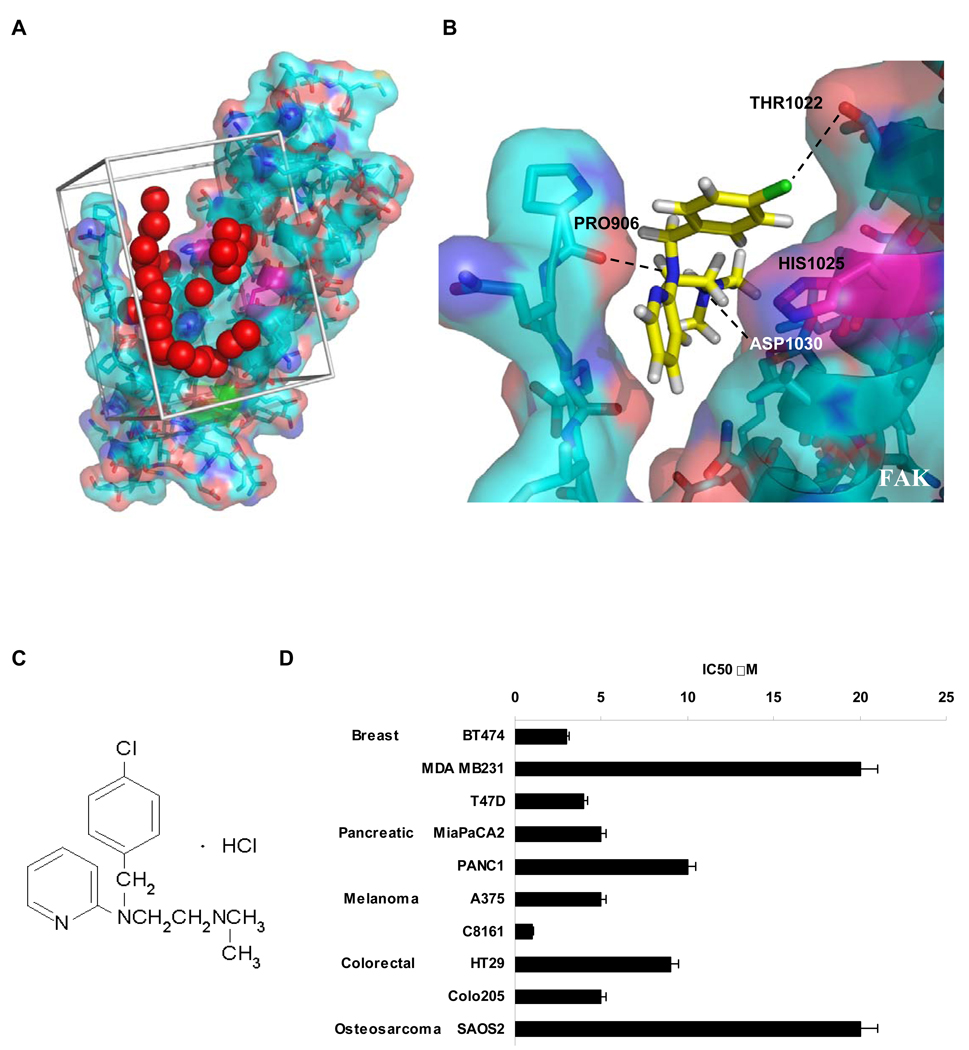
Figure 1.
Structure-based development of small molecules that targeted the binding of FAK and VEGFR-3. A, Site selection for high throughout virtual screening of drug-like compounds to develop small molecule FAK inhibitors. The crystal structure of the Focal Adhesion Targeting (FAT) domain of FAK was obtained from the Protein Data Bank (PDB ID: 1K04), and prepared for computational docking. 140,000 small molecules from the NCI’s Development Therapeutics Program were each positioned in the structural pocket and scored for electrostatic and van der Waals interactions as implemented in DOCKv5.2 package9 (UCSF). The crystal structure is shown in cyan and salmon, and residues that undergo shifts upon peptide binding in NMR studies are shown in magenta. The catalytic tyrosine is shown in green. Red spheres indicate the site defined by the program SPHGEN (UCSF) with chemical and geometric features appropriate for specific small molecule binding. Grey bars demarcate the scoring grid utilized to calculate interactions between potential ligands and the targeted structural pocket. B, Predicted binding site of 1 to Focal Adhesion Targeting domain of FAK. Histidine 1025 shown in magenta with surrounding residues. Black dashed lines indicate hydrogen bonds between Proline 906 and 1 N2, Aspartic acid 1030 hydroxyl group and 1 N1 and also hydroxyl group of Threonin1022 and Cl of 1.
C, Compound 1 (Chlorpyramin hydrochloride) structure.
D, 1 treatment decreased the viability of a diverse set of cancer cell types. MTS assay was performed on selected cell lines. Cells were treated with the increased concentration of 1 for 72 h and analyzed with Cell Titer Proliferation Assay; error bars represent ±SEM. P < 0.05.