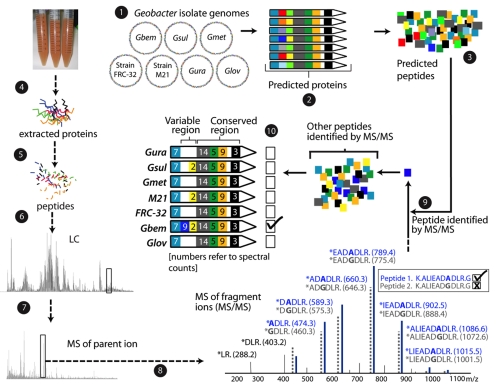
FIG. 1.
Strain-resolved proteogenomic techniques allow identification of unique peptides and constrain the genotypes present in environmental samples. (1) Proteins were predicted from the seven isolate Geobacter genomes. (2) Proteins were aligned. In the example shown, they share a conserved region (similar colors in all seven regions) and have a variable region (indicated by the variety of colors for each protein). (3) Tryptic digest patterns are used to predict peptide sequences. Proteins were extracted from the experimental sample (4) and digested into peptides by using trypsin (5). These peptides were separated using LC (6) before high-resolution mass spectrometry on a fraction of peptides from an elution peak (7) to determine the mass of the parent ions. (8) The peptides are fragmented and mass spectrometry measurements made on fragment ions. (9) Mass spectral data are matched to peptide sequences predicted in step 3 to allow peptide identification. Peptide sequences differing by only one amino acid can be identified, as shown in the spectral diagram. (10) Detected peptides are mapped onto aligned protein sequences to identify peptides shared or unique to each of the isolate species. Spectral counts can be mapped together with peptides. Where coexisting unique peptides are detected, ratios of unique spectral counts can be used to infer relative strain abundance. As shown, the identification of a unique peptide indicates that this protein is most closely related to that of G. bemidjiensis.