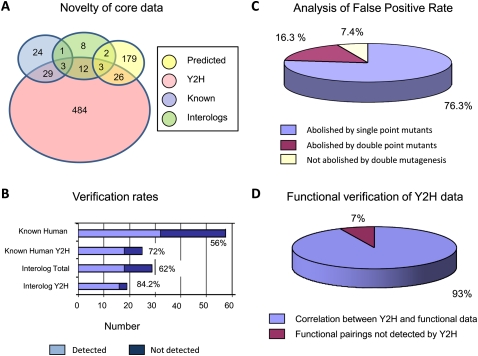
Figure 2.
Analysis of data from targeted Y2H experiments. (A) The novelty of Y2H data was determined relative to known interactions contained within the MINT, IntAct, BioGRID, and HPRD databases, known Interolog interactions, or non-Interolog predicted interactions (derived from the Hi-map or IntNet databases). (B) Reconfirmation of known or predicted E2/E3-RING interactions is highly dependent upon the source of the predicted data. (C) Strategic mutagenesis studies were performed to establish the proportion of positive Y2H interactions that conform to the known molecular/structural requirements for E2/E3-RING complex formation (see experimental data presented in Supplemental File 5). (D) To assess the efficiency with which Y2H data can predict functionally active E2/E3-RING complexes, 51 different E2/E3-RING combinations were systematically tested for ubiquitination activity in vitro (see experimental data presented in Supplemental File 6). Activity profiles for specific E2/E3-RING complexes were then compared with Y2H data to establish the percentage correlation between the two data sets.