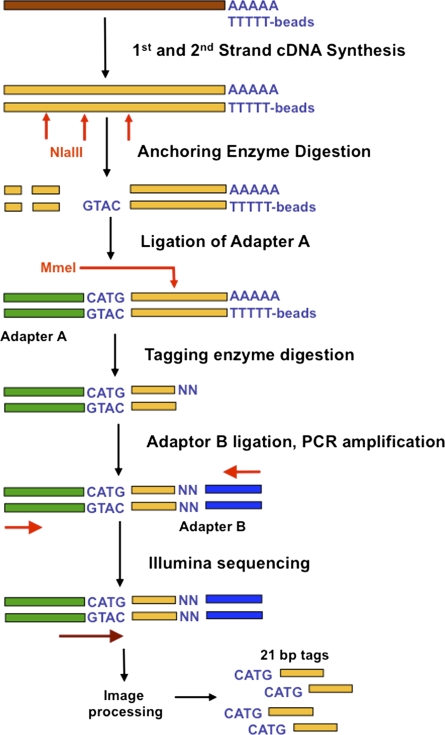
Figure 1.
Outline of Tag-seq library generation. Each mRNA (brown) underwent double-stranded cDNA synthesis using oligo(dT) beads, to capture polyadenylated RNA. cDNA (gold) is digested with the NlaIII anchoring restriction enzyme (vertical red arrows), leaving a 4-bp overhang (GTAC). Only cDNA fragments anchored to oligo(dT) beads are retained. Adapter A (green) is ligated to the overhang and adds a recognition site for the TypeIIS tagging enzyme MmeI. Following MmeI digestion (red vertical arrow), a second adapter is ligated (Adapter B, blue) to the resulting 2-bp overhang. PCR primers (horizontal red arrows) annealing to adapters A and B are used to enrich tags. Cluster generation and sequencing (horizontal brown arrow) is performed on the Illumina cluster station and analyzer. The resulting image files are processed to extract the read sequences, and 21-bp SAGE tags are further extracted from the reads. Tags consist of the 4-bp NlaIII recognition sites and 17 bp of unique sequence, and constitute a total of 21 bases that can be mapped back to the original mRNA (brown).